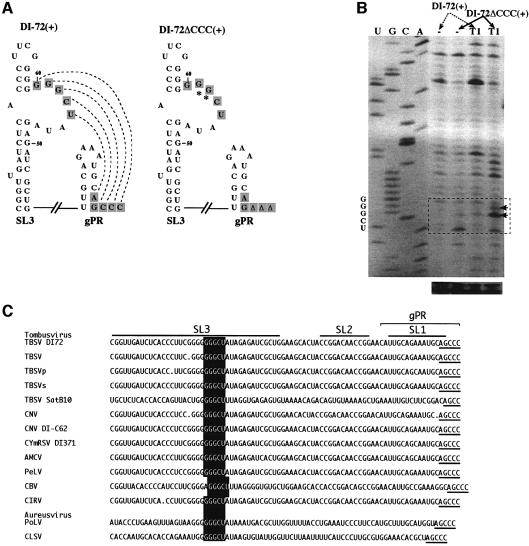
Fig. 4. An RNA–RNA interaction between gPR and SL3 is supported by enzymatic probing of DI-72(+). (A) Schematic representation of the putative interaction, indicated with dotted lines, between the interior loop in SL3 and the 3′ end in DI-72(+), and the lack of interaction in DI-72ΔCCC(+).The predicted secondary structures of gPR and SL3 are shown with complementary sequences boxed. The RNase T1 (T1) sensitive nucleotides in the internal loop are indicated with arrows (see below). (B) Enzymatic probing of DI-72(+) and DI-72ΔCCC(+). The in vitro-transcribed RNAs were treated in vitro with T1 or not treated (control), followed by analysis by reverse transcription-based primer extension using primer PMF59S in the presence of [35S]dATP, and separated on an 8% sequencing gel. The areas boxed with dotted lines include the interior-loop region of SL3. (C) Sequence comparison of the replication silencer region and the 3′ ends in tombusviruses. Sequences are shown in the 5′-to-3′direction, with AGCCC as the 3′ end. The predicted secondary structure for DI-72 is shown schematically on the top. The conserved silencer sequence is boxed, while the complementary 3′ terminal pentanucleotide is underlined.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.