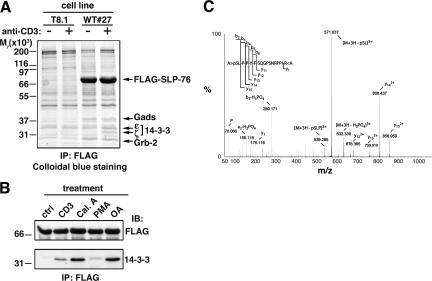
Figure 1.
Detection of inducible association of 14-3-3 proteins to SLP-76 and serine 376 phosphorylation. (A) WT#27 cells overexpressing FLAG–SLP-76 and parental T8.1 cells were left unstimulated (−) or activated by anti-CD3 cross-linking (+) for 5 min at 37°C. Lysates were incubated with anti-FLAG affinity resin, and bound proteins were eluted with an excess of FLAG peptide. Eluates were then fractionated by SDS-PAGE, and proteins were detected by colloidal blue staining. Relevant protein bands were excised, digested in gel by trypins, and identified by MALDI-TOF peptide mass fingerprinting. Migration of protein standards is indicated on the left. (B) WT#27 cells were left unstimulated (ctrl), activated at 37°C by CD3 cross-linking as in A, or treated with 1 μM calyculin A (Cal.A), 10 ng/ml PMA, or 1 μM okadaic acid (OA) for 30 min at 37°C. Lysates were immunoprecipitated with anti-FLAG antibodies as described in A and immunoblotted with anti-FLAG (top) or anti–14-3-3 (bottom) antibodies. (C) FLAG–SLP-76 was immunoprecipitated from CD3 cross-linking–stimulated WT#27 cells as described in A. The protein band was excised and digested in gel by trypsin and AspN. The resulting peptide mixture was analyzed by nanoESI MS. The acquired MS/MS dataset was submitted to sequence tag scanning to spot modified peptides. The MS/MS spectrum shows fragment ions of a peptide spanning residues 376–391 of SLP-76, which clearly demonstrates phosphorylation of S376.