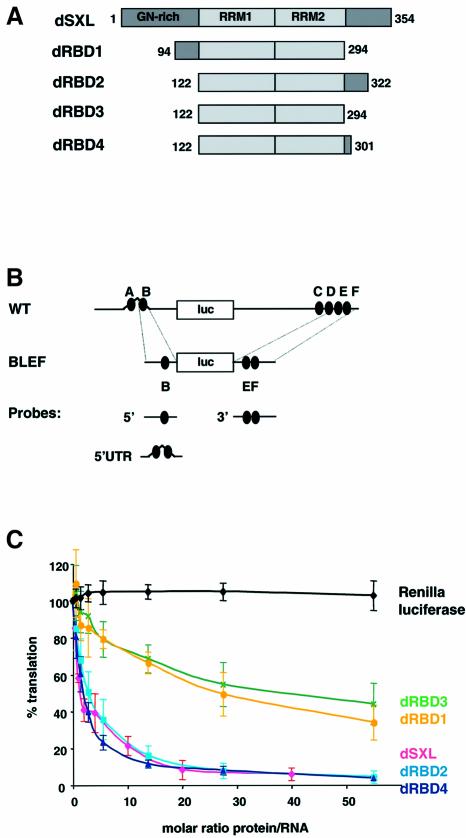
Fig. 1. Mapping the translational repressor domain of dSXL. (A) Schematic representation and domain organization of dSXL and its deletion derivatives. The amino acid numbers included in each derivative are indicated. (B) Scheme of the RNA constructs used in this study. WT mRNA contains the full-length 5′ (626 nt) and 3′ (1047 nt) UTRs of msl-2 fused to the firefly luciferase open reading frame (Gebauer et al., 1999). The SXL-binding sites are denoted A to F (black ovals). BLEF mRNA contains the minimal msl-2 sequences required for translational repression, which consist of 69 nt in the 5′UTR including site B, and 46 nt in the 3′UTR including sites E and F (Gebauer et al., 2003). Probes used for gel mobility-shift and UV-crosslink assays are also depicted. (C) WT mRNA was translated in Drosophila embryo extracts in the presence of increasing amounts of dSXL (pink line), dRBD1 (yellow line), dRBD2 (light blue line), dRBD3 (green line) or dRBD4 (dark blue line). Renilla luciferase mRNA was co-translated as an internal control (black line). Firefly luciferase values were corrected for Renilla expression and plotted as the percentage of the activity obtained in the absence of recombinant protein against the molar ratio of protein to mRNA.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.