Figure 1.
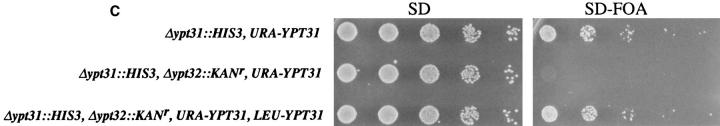
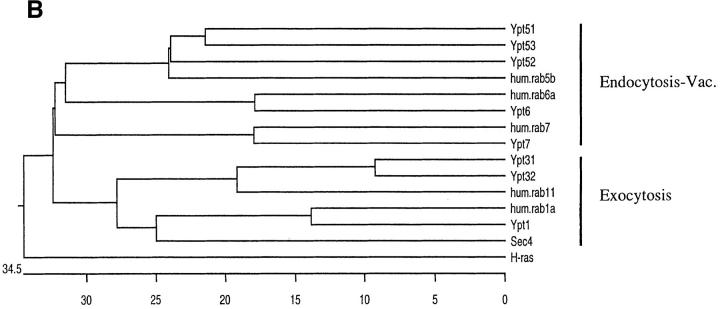
The YPT31 and YPT32 genes encode two functionally homologous exocytic GTPases. (A) Comparison of the amino acid sequences of Ypt31 and Ypt32 proteins. The predicted protein sequence of Ypt32 was compared to Ypt31 using the MegAlign program (DNAStar Inc., Madison, WI, Clustal method with the PAM250 residue weight table). Identities are shaded with solid black, and residues conserved to within two distance units are shaded. Overall, the two proteins are 81.1% identical and 89.6% similar when compared using the bestfit program (Genetics Computer Group, Madison, WI). (B) Ypt31 and Ypt32 proteins belong to a subfamily of exocytic Ypt GTPases by phylogenetic analysis of the Ypt/rab family of small GTPases. The predicted amino acid sequence of Ypt31 and Ypt32 were compared to all other Ypt proteins, using the completed S. cerevisiae genome sequence and their human homologues. The analysis shows that Ypt/rab proteins fall into two functional subfamilies: those involved in endocytosis and vacuolar protein sorting (Endocytosis-Vac.) and those involved in exocytosis. Sequences were aligned as above. The scale at the bottom indicates the number of substitutions between sequences. hum., Homo sapiens; H-ras, Harvey murine sarcoma virus ras protein; all other sequences, Saccharomyces cerevisiae. These sequence data are available from GenBank/EMBL/DDBJ under accession numbers: Ypt31, U18778; Ypt32, X72834; rab11, X56740; Ypt7, X68144; rab7, U44104; Ypt1, X00209; rab1a, M28209; Sec4, M16507; Ypt6, U17244; rab6a, M28212; Ypt51, X76173; Ypt52, X76174; Ypt53, X76175; rab5b, X54871; and H-ras, X00740. (C) Requirement of YPT31 or YPT32 genes for cell viability. The YPT31 gene was precisely deleted using the HIS3 gene, and this strain was transformed with a URA3-marked CEN vector containing the YPT31 gene under control of its own promoter (NSY301, first row). This strain (NSY301) was subsequently deleted for the YPT32 gene using the KANr gene as a dominant delectable marker (NSY302, middle row). Finally, this strain (NSY302) was transformed with a second plasmid marked with LEU2 and carrying the YPT31 gene (NSY306, bottom row). The three strains were grown in synthetic media maintaining selection for plasmids. Serial dilutions of cells were then spotted onto either SD or SD-FOA and grown at 26°C. Cells deleted for both genes do not grow on SD-FOA plates, indicating that they cannot lose the URA-marked YPT31 plasmid unless they carry also the LEU-marked YPT31 plasmid.