Abstract
Genes with unknown function are called orphan genes while their transcripts and peptides are called hypothetical proteins. There are many genes and their associated proteins that remain uncharacterized in the human genome. A database of human hypothetical proteins with ascribed functions could be helpful for biologists to search for potential proteins of interest. In recent years, the rapid completion of genome sequences has created essential information to link genes to gene products. In order to better explain functions for un-annotated proteins we designed BioinformaTRICKS (an open source project) and used it to develop a database called HYPO.
Availability
The database is available for free at http://pc-dugong.ruc.dk:8080.
Keywords: hypothetical proteins, mining, database, networks
Background
As the amount of genome sequence data now available is enormous with more than 750 genomes being either finished or in progress, a biologist is thrown into using several databases with increasing attention to find any novel genes or proteins or function. However, various analyses based on sequence, structure, function and “Omic” data have revealed different annotation criteria leading to different sets of predicted genes. However, more than 50% of proteins in the proteome zone remain un-annotated and un-identified for function (Table 1 in supplementary material). The human genome contains many different regulatory sequences that have roles in controlling gene expression. The protein-coding sequences is only less than 1.5 percent of the genome and then rest remains as non-coding inter and intra-genic regions with undetermined role. [1]
The annotation of various human chromosomes is well supported by computational predictions where there is no similarity to known proteins or EST sequences. The genes that have unknown function called as orphan genes code for proteins annotated as “hypothetical proteins”. Hence, there is a need to begin constructing and analyzing protein families clustered as “hypothetical proteins” with an aim to elucidate function and protein subunit interactions.
After several drafts of Human Genome Project, there are many proteins that remain to be characterized. Tools have been developed to utilize evolutionary relationships towards understanding uncharacterized proteins despite the need to generate functional interaction networks. [2,3] In particular, these approaches are being used to annotate functions for hypothetical proteins. Although several databases explore protein functions through data-mining, there is a requirement to list all hypothetical proteins. There are reports that address the problem of orphan genes. [4] However, there is no adequate information to necessitate function of genes that cannot be based on homology alone, except connected to other known gene family.
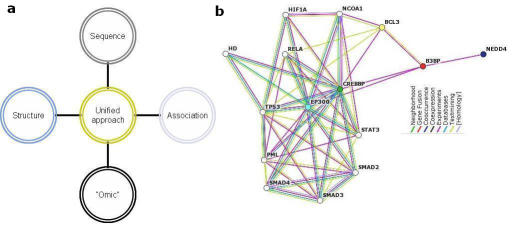
There are several hypothetical proteins such as the KIAA that have remained hype for some time now. [5] Systems biology integrated with protein-protein interaction (PPI) studies could identify the role of these unknown proteins (Figure 1a). Systems biology is a science of constructing networks of genes and proteins thereby providing a framework for predicting models. [6] The proteins connected through the networks could perhaps throw light on the plausible function of the hypothetical proteins through the bona fide interactions they are involved with.
Figure 1.
(a) Integrating four methods in making up a database of hypothetical proteins viz., sequence based, structure based, association based and omic based. All the protein accessions were mined using a query (keywords hypothetical AND Homo sapiens) from NCBI searches (www.ncbi.nlm.nih.gov); for complete list of hypothetical proteins, please visit http://pc-dugong. ruc.dk:8080). (b) Interactors of B3BP are associated through different approaches (Please find the legend in different colors for different approaches). The network neighbourhood of B3BP (Marked in red; Accession #NP_438169) is shown as hypothetical when Entrez NCBI query is made. When this accession was queried using STRING, it facilitates in understanding how and what all the other proteins interacting with NP_438169 are doing in a network
In the context of PPI networks, we could consider if a model is to be developed from the network or a network is to be generated with an already established model. Precisely, the putative function of a protein could be better known from a PPI network to develop a model from it. We show here an example in finding the putative function of a hypothetical protein (figure 1b, NP_438169 - B3BP Homo sapiens Nedd4-binding protein 2) using a PPI network. The nearest interacting partners of the protein B3BP were mapped using the STRING that could show the probable function of the query [7]. Information on “known” or “unknown” protein-protein interactions is still mostly limited but integrating tools such as these could generalize a way to find the function of hypothetical proteins.
While we started mining the proteins, it seemed that there are a few hypothetical proteins that have amino acid residues HYP (histidine, tyrosine and proline) in succession. These might have been long-established through the mutations that are introduced into the proteins at one or more predicted non-essential residues.
While a few KIAA proteins are conserved and have been known to be identified as novel [8], functional analysis of the proteins encoded by these KIAA cDNAs could be established from our database of hypothetical proteins. [5] A “conservative amino acid substitution” is the one in which the amino acid residue is replaced with an amino acid residue having a similar side chain. The families of amino acid residues having similar side chains are distinct and include conserved amino acids such as histidine (aromatic/basic side chains), tyrosine (polar side chain) and proline (non polar side chains). On the other hand, these might have appeared during annotation through the mutations introduced randomly along all or part of a coding sequence. Our database has over 6362 hypothetical proteins that could be searched for different functions.
Observations and challenges
We observe in tandem that few hypothetical proteins present on different chromosomal loci are known to have the same putative function. Categorizing several approaches beyond traditional sequence similarity that utilize tremendously large amounts of data that is available for computational prediction of functions is the need of the hour these days. Having said this, one could use a subset of proteins that match from several of the experimental approaches and be used as a predictor to circumvent the use of wet laboratory experiments in the near future.
Another concern now is specifically on hypothetical protein domains in intrinsically disordered proteins (IDPs). [7] With Protein Data Bank (PDB), not accepting theoretical structures, there is an emphasis for experimental approach by researchers to determine the structure and functional relationship of a protein. Nevertheless, carefully considering the functional annotation methods as discussed above could definitely devise selection for proteins that could be experimented. This could be an interesting approach to pursue further.
Yet another issue to be noted is the appraisal to understand if any of the hypothetical proteins have proper functional annotations’ been attributed to sequence: structure: function relationship in case of ordered proteins while sequence: un-structure: function in case of intrinsically disordered proteins. In conclusion, the current methods could play an important role in establishing functions for proteins annotated as hypothetical in the genome.
Note
The title of the article contains hypo abbreviated for hypothetical proteins.
Supplementary material
Acknowledgments
Thanks are due to the following who have contributed in mining the proteins: Renuka Suravajhala, Roskilde University, Denmark; Tarun Gupta, Panjab University, Chandigarh, India, Umesh Roy, Precision Biotech, New Delhi, India; Chandan Badapanda, National Chemical Laboratory, Pune, India; Arun Gupta, Satyabama University, Chennai, India; Shirish Siddamshetty, St. Martin's college, Hyderabad, India. This project is hosted in bioinformatics.org and bioclues.org. All the contributors are graduate students working virtually for a non-profit bioinformatics community: bioclues.org.
References
- 1.Little PF. Genome Res. 2005;15:1759. doi: 10.1101/gr.4560905. [DOI] [PubMed] [Google Scholar]
- 2.Yamasaki C, et al. Gene. 2005;364:99. doi: 10.1016/j.gene.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 3.Chen F, et al. Nucleic Acids Research. 2006;34:D363. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blayo P, et al. Theoretical Computer Science. 2003;290:1407. [Google Scholar]
- 5.Nagase T, et al. Brief Funct Genomic Proteomic. 2006;5:4. doi: 10.1093/bfgp/ell005. [DOI] [PubMed] [Google Scholar]
- 6.Evanko D. Nat Methods. 2006;3:964. doi: 10.1038/nmeth1206-964b. [DOI] [PubMed] [Google Scholar]
- 7.Radivojac P, et al. Biophys J. 2007;92:1439. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doerr A. Nat Methods. 2007;4:8. doi: 10.1038/nmeth0107-8b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.