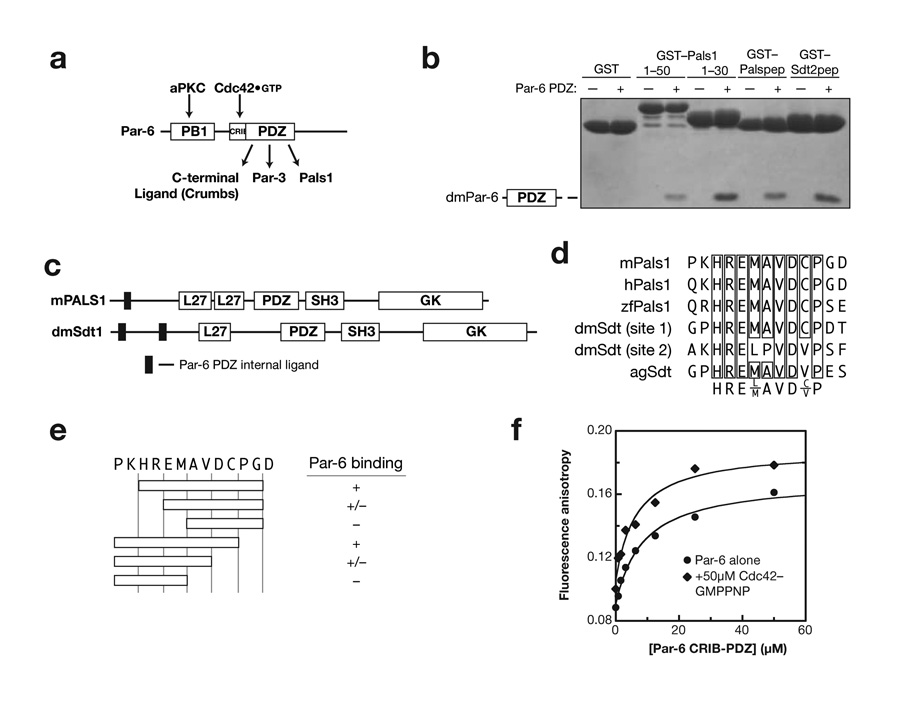
Figure 1.
Identification of Par-6 PDZ binding motifs in Pals1 and Stardust. (a) Schematic of Par-6 domain structure and domain ligands (PB1: Phox and Bem1; CRIB: Cdc42/Rac Interactive Binding; PDZ: PSD-95, Discs Large, ZO-1). (b) Deletion analysis of Pals1 and Sdt reveals a short, internal Par-6 PDZ ligand. GST fusions of various murine Pals1 or Drosophila Stardust fragments were tested for their ability to bind Par-6 using glutathione agarose affinity chromatography (Palspep: Pals1 residues 29 to 40; Sdt2pep: Sdt residues 375 to 388). (c) Domain structure of Pals1 and Sdt showing the relationship of the Par-6 PDZ internal ligands to the MAGUK protein domains (L27: Lin-2, Lin-7; SH3: Src Homology 3; GK: Guanylate Kinase Homology). (d) Alignment of Pals1/Sdt Par-6 ligand sequences showing conserved core sequence. (e) Deletion analysis of Pals1/Sdt Par-6 ligand sequences. The conserved core sequence is required for binding to the Par-6 PDZ domain. (f) The Rho GTPase Cdc42 does not regulate Pals1 binding to the Par-6 PDZ domain. A rhodamine labeled peptide with the Pals1 sequence was used to measure binding to the Par-6 CRIB-PDZ fragment. The higher anisotropy of the Cdc42 complex at saturation occurs because of its higher molecular weight. The binding curves are for Kd’s of 6 and 8 μM for Cdc42-bound and free Par-6, respectively.