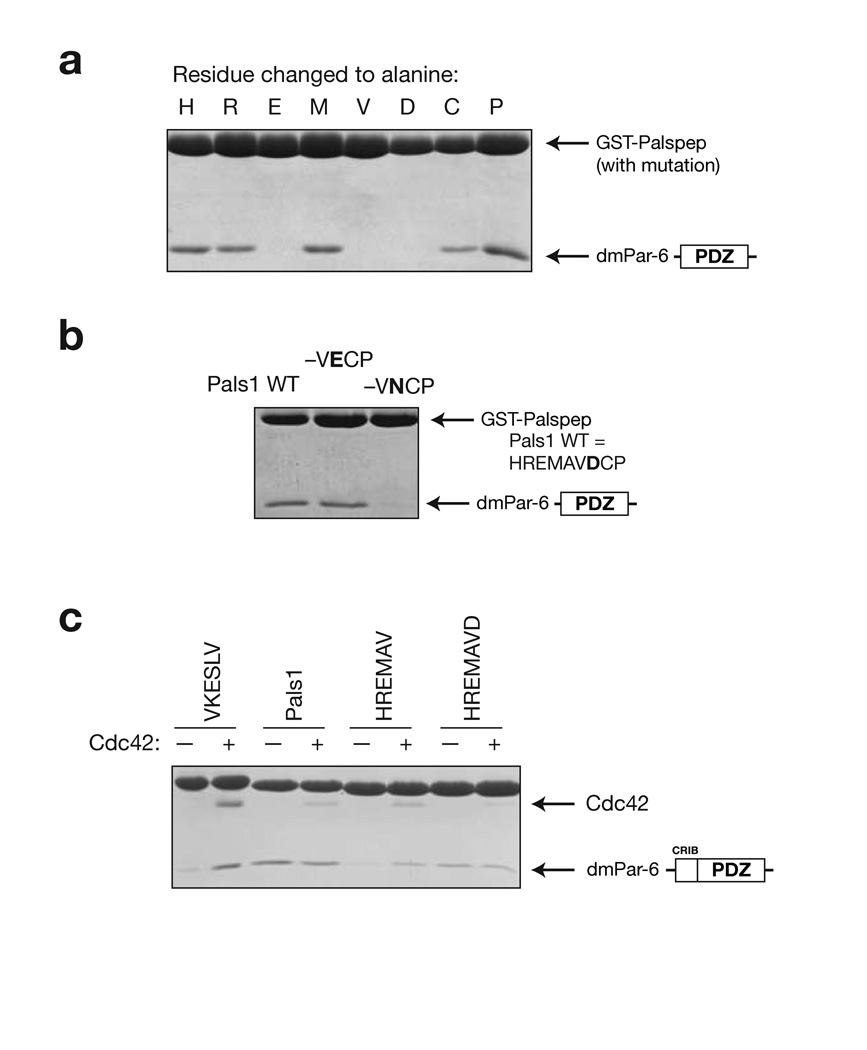
Figure 4.
Analysis of Par-6 PDZ ligand binding determinants. (a) Alanine scanning of Pals1 ligand. GST-fusions of peptide sequences with an alanine substituted at each position along the Pals1 sequence were tested for their ability to bind the Par-6 PDZ domain. (b) Evaluation of the importance of the P+1 residue in Pals1. In the structure of the Par-6 Pals1 complex, the P+1 aspartic acid forms a salt bridge with a conserved lysine in the PDZ domain. GST fusions of mutants in this position were tested for their ability to bind the Par-6 PDZ domain. Mutation to an asparagine abrogates binding indicating that the salt bridge is critical for binding. (c) Identifying the determinants of Cdc42 regulation. As has been previously shown, Cdc42 binding to the CRIB-PDZ fragment of Par-6 alters its affinity for carboxy terminal ligands. GST-fusions of several Pals1 truncations were made to identify the determinants of Cdc42 regulation. When the P+1 residue is removed (HREMAV sequence), Cdc42 binding causes an increase in peptide binding. The presence of this residue (HREMAVD sequence) causes Cdc42 to no longer affect peptide binding, presumably because the carboxylate-binding loop is deformed.