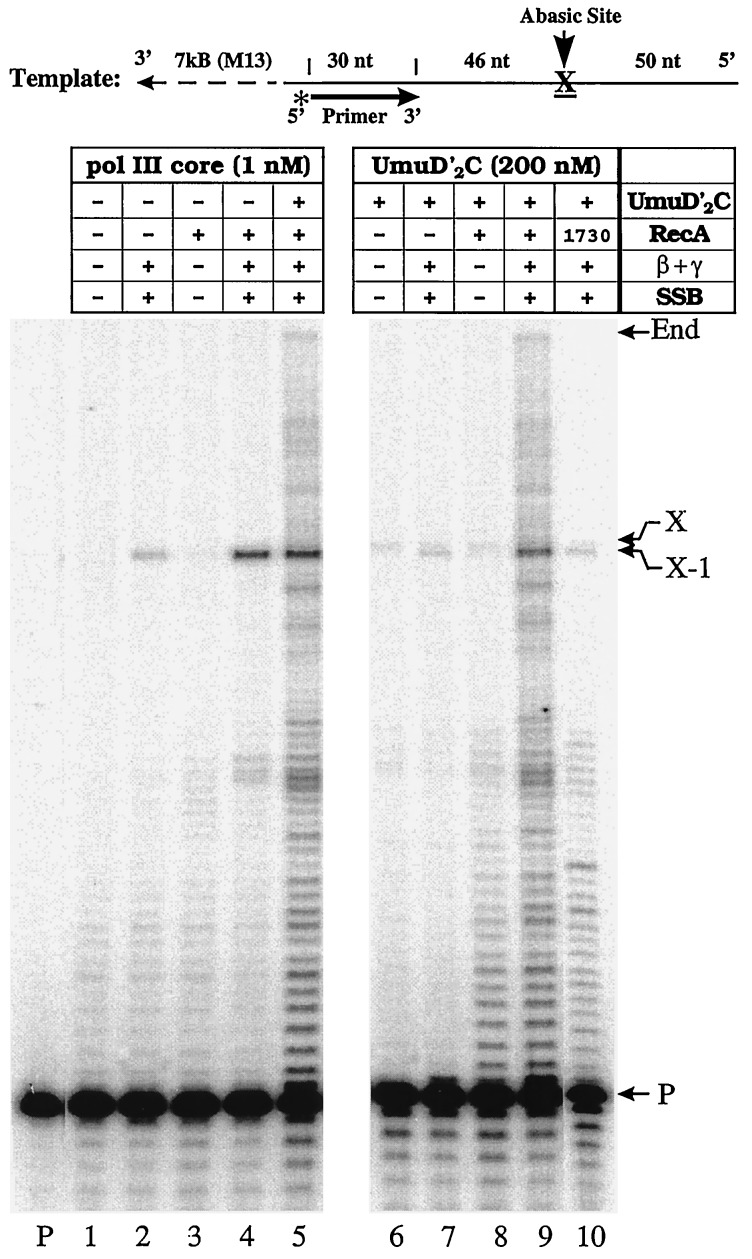
Figure 1.
UmuD′2C-stimulated abasic site bypass. Standard polymerization reactions, using a running-start protocol, were carried out in the presence or absence of exogenous pol III core by using combinations of UmuD′2C, RecA, β,γ-complex, and SSB. Four dNTPs (100 μM) and ATP (1 mM) were present in all reactions. A 32P-labeled primer was annealed to a DNA template containing an abasic lesion, X (top of figure), and the replication products were separated in 10% denaturing polyacrylamide gels and visualized by phosphorimaging. Locations of the unextended primer band, abasic site (X), upstream site adjacent to the lesion (X − 1), and the end of template are indicated on the right. Lane P contains the primer in the absence of proteins. Additions to the replication reaction mixtures are shown in the box at the top of the gel; β + γ, represents the β-clamp processivity subunit of the pol III holoenzyme complex and the five protein γ-clamp loading complex consisting of the subunits γ, δ, δ′, χ, ψ.