Figure 1.
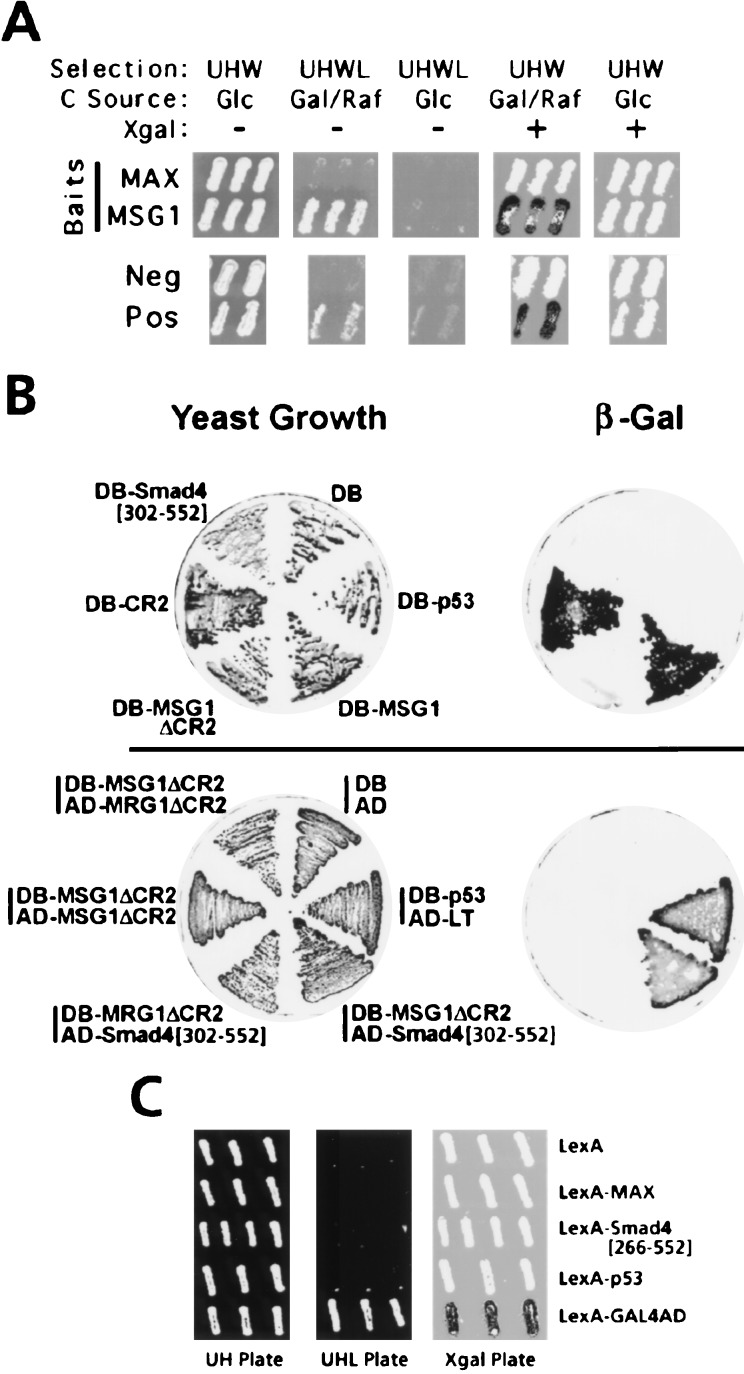
Functional interaction and transcriptional-activating activities of MSG1 and Smad4(C) in yeast. (A) Functional interaction of MSG1 and Smad4(C) demonstrated by the interaction-trap yeast two-hybrid assay (19, 27). Smad4(C) prey is expressed on galactose/raffinose (Gal/Raf) plates but not on glucose (Glc) plates (19, 27). Positive (Pos) and negative (Neg) control interactors and the LexA-Max bait (MAX, nonspecific interactor control) are described (27). Selection markers were used for: β-galactosidase reporter plasmid (Ura+, U); LexA-fusion bait plasmids (His+, H); B16 activator-fusion prey plasmids (Trp+, W); and the auxotrophic phenotype resulting from interaction of the bait and prey (Leu+; L). (B) Functional interaction and transactivating activities of MSG1, MRG1, and Smad4(C) analyzed by GAL4-based yeast one-hybrid (Upper) or two-hybrid (Lower) assays (20). DB, GAL4 DNA-binding domain; AD, GAL4 transcriptional activating domain; LT, simian virus 40 large T antigen. Transcription of the β-galactosidase reporter gene by transactivating activity of GAL4DB fusion proteins (Upper) or resulting from protein–protein interaction GAL4DB- and GAL4AD-fusion proteins (Lower) were evaluated by the β-galactosidase replica filter assay (15). (C) Absence of transactivation by Smad4[266–552] in yeast. Transactivating activities of LexA-fusion proteins (His+ selection, H) were evaluated as described (19), by the LexA-dependent Leu+ (L) auxotrophic phenotype (determined on the UHL plate) or transcription of a LexA-dependent β-galactosidase reporter plasmid (Ura+ selection, U) (determined on the Xgal plate). The carbon source of all plates were galactose/raffinose. A fusion protein of LexA and GAL4 transactivating domain (GAL4AD) was used as positive control.