Abstract
The Drosophila myoblast city (mbc) locus was previously identified on the basis of a defect in myoblast fusion (Rushton et al., 1995. Development [Camb.]. 121:1979–1988). We describe herein the isolation and characterization of the mbc gene. The mbc transcript and its encoded protein are expressed in a broad range of tissues, including somatic myoblasts, cardial cells, and visceral mesoderm. It is also expressed in the pole cells and in ectodermally derived tissues, including the epidermis. Consistent with this latter expression, mbc mutant embryos exhibit defects in dorsal closure and cytoskeletal organization in the migrating epidermis. Both the mesodermal and ectodermal defects are reminiscent of those induced by altered forms of Drac1 and suggest that mbc may function in the same pathway. MBC bears striking homology to human DOCK180, which interacts with the SH2-SH3 adapter protein Crk and may play a role in signal transduction from focal adhesions. Taken together, these results suggest the possibility that MBC is an intermediate in a signal transduction pathway from the rho/rac family of GTPases to events in the cytoskeleton and that this pathway may be used during myoblast fusion and dorsal closure.
In vertebrate organisms, a common feature of the myogenic differentiation program of all muscle fibers is the apparent recognition, adherence, and fusion between myoblasts that generate multinucleate syncitia (for review see Fischman, 1972; Wakelam, 1985). Since this process can occur in cultured cells, tissue culture systems have been invaluable in identifying regulators of myoblast fusion. Essential components include cell adhesion molecules, calcium and molecules that are regulated by it, metalloproteases, meltrins, lipids, and others (Yagami-Hiromasa et al., 1995; for reviews see Wakelam, 1985; Knudsen, 1991). Homologues to these vertebrate factors have not yet been shown to function in myogenesis in Drosophila. However, morphological studies have established that the differentiated muscle fibers of insects are also syncitial (Ball et al., 1985; Campos-Ortega and Hartenstein, 1985; Bate, 1990, 1993; Doberstein et al., 1997). Therefore, one might anticipate that similar molecules will control fundamental aspects of myoblast fusion in a variety of species. Moreover, the use of genetics to identify critical myogenic regulators and their relationship to other molecules in Drosophila seems likely to reveal parallel pathways in vertebrate organisms.
While it seems unlikely that all aspects of myogenesis will be analogous between Drosophila and vertebrates, many appear to be conserved between these organisms. One example is the apparent conservation of myogenic regulatory molecules. Among these are nautilus (Michelson et al., 1990; Paterson et al., 1991), which, like its vertebrate counterparts (for review see Weintraub, 1993), can induce a somatic muscle differentiation program (Keller et al., 1997) and MEF2, an enhancer binding protein that is absolutely required for the induction of muscle-specific structural genes and myogenic differentiation (Lin et al., 1996; for review see Olson et al., 1995). Similarities are also apparent at the cellular level. For example, extensive proliferation of myosin-expressing myoblasts is not observed in either vertebrates (for review see Holtzer et al., 1975a ) or Drosophila (Campos-Ortega and Hartenstein, 1985; Bate, 1993; Rushton et al., 1995). In addition, although fusion normally occurs before myosin expression in vertebrate cells, it is not an absolute prerequisite for expression of myosin in either system (Holtzer et al., 1975a , and references therein; see also Emerson and Beckner, 1975; Endo and Nadal-Ginard, 1987; Luo et al., 1994; Paululat et al., 1995; Rushton et al., 1995; Doberstein et al., 1997). Finally, recent studies have revealed striking similarities between the ultrastructure of fusing Drosophila myoblasts (Doberstein et al., 1997) and fusing vertebrate myoblasts (Engel et al., 1985; for review see Kalderon, 1980).
The above studies suggest several parallels between myogenesis in Drosophila and in vertebrate systems. However, little is actually known about the genes regulating myoblast fusion in Drosophila and their mechanism of action. Recently, several loss-of-function mutations that exhibit defects in myoblast fusion have been identified, including rolling stone (Paululat et al., 1995), myoblast city (mbc)1 (Rushton et al., 1995), blown fuse (Doberstein et al., 1997), sticks and stones (Abmayr, S.M., M.R. Erickson, B.A. Bour, and M. Kulp. J. Cell. Biochem. 1994. 18D (Suppl.):474), and singles bar (Maeland, A.D., J.W. Bloor, and N.H. Brown. 1996. Mol. Biol. Cell. 7:39A). The protein coding sequence of blown fuse, the first of these genes to be identified, has not yet provided insight into its function. By comparison, examination of altered forms of the small rho-like GTPase, Drac1, has been useful in understanding myoblast fusion in Drosophila (Luo et al., 1994). Drac1 is the Drosophila homologue of the vertebrate gene rac1, which has been shown to induce membrane ruffling through reorganization of the actin cytoskeleton (Ridley et al., 1992). Both dominant negative and constitutively active forms of Drac1 have been shown to cause defects in myoblast fusion (Luo et al., 1994). Interestingly, altered forms of Drac1 also disrupt the actin cytoskeleton in the epidermis, causing defects in cell migration and dorsal closure (Harden et al., 1995), and in apical regions of the wing imaginal disc (Eaton et al., 1995).
A role for the cytoskeleton in myoblast fusion in vertebrates has previously been shown using low concentrations of the inhibitor cytochalasin B, which interferes with the formation of actin filaments. In these studies, the fusion of myoblasts in culture was severely limited in the presence of cytochalasin B, and most myotubes contained only two nuclei (Sanger et al., 1971; Sanger and Holtzer, 1972). More recent studies have confirmed that both cytochalasin B and D inhibit myoblast fusion and correlate the lack of fusion with the disruption of actin filaments (Constantin et al., 1995). While the role of the cytoskeleton at this early stage of myoblast fusion remains unclear, it may be related to the formation of lipid-rich domains within the cell membrane. Just before fusion, for example, vertebrate myoblasts have been shown to undergo a topological change that results in the creation of protein-depleted, lipid-enriched membrane domains (Kalderon and Gilula, 1979; Fulton et al., 1981). These lipid-rich domains are believed to be associated with an increase in membrane fluidity (for review see Wakelam, 1985) and may create sites for membrane–membrane fusion. Thus, subcellular structures that organize these lipid-rich domains may be dependent on cytoskeletal rearrangements.
Herein we describe the isolation and characterization of the mbc gene. MBC is one of the first proteins identified in Drosophila that is essential for myoblast fusion. It is expressed in a broad range of tissues throughout embryonic development, including the presumptive musculature and epidermal cells involved in the process of dorsal closure. Consistent with its expression pattern, mbc mutant embryos exhibit defects in dorsal closure and cytoskeletal organization as well as myoblast fusion. These abnormalities are similar to those described above for the small GTPase Drac1, and suggest that (a) mbc functions in the same pathway as Drac1 in the epidermis and (b) this pathway is used in the mesoderm for events leading to myoblast fusion. MBC has striking homology to DOCK180, a human gene that was identified on the basis of interaction with the small adapter protein Crk. DOCK180 may be involved in signal transduction from focal adhesions, and results reported herein are consistent with a similar function for MBC. Finally, open reading frames (ORFs) from several genome projects suggest that DOCK180 and MBC define a new gene family.
Materials and Methods
Drosophila Stocks
All stocks were grown on standard cornmeal medium at 18 or 25°C, as necessary. Balancer and marked chromosomes are described in FlyBase (http://cbbridges.harvard.edu:7081). Df(3R)mbc-30 has been described (Rushton et al., 1995). Df(3R)mbc-15A was created by treating males homozygous for P{ry +, lacZ}A189.2F3 (Bloomington Stock Center) with 4,000 rads of γ-rays. Approximately 25,000 chromosomes were screened for the loss of the ry + marker. Three deficiencies, including Df(3R)mbc-15A, were recovered.
Df(3R)CA15 and Df(3R)CA2 were obtained by imprecise excision of the homozygous lethal P-element insertion l(3)04684 (Bloomington Stock Center, Bloomington, IN). Mobilization occurred in flies carrying l(3)04684 over Sb, Delta2-3 ry, and excision events were recovered over MKRS or TM2. 677 excision events were analyzed. 330 imprecise excision events were identified by lack of complementation of Df(3R)mbc-F5.3/ TM3 (see Fig. 1). These were subsequently reevaluated for lack of complementation of l(3)95BCd and l(3)01152 and complementation of mbc S4. Four deficiencies were obtained, two of which are shown in Fig. 1.
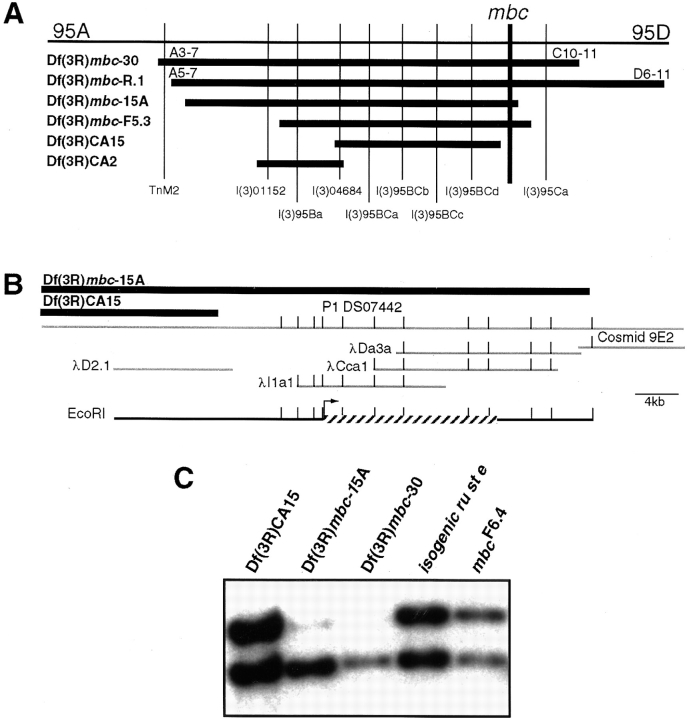
Figure 1.
Genetic and molecular map of cytological region 95BC. (A) Deficiencies are represented by horizontal bars and lethal complementation groups by vertical lines. Groups l(3)95BCa-d have not been oriented with respect to each other. (B) Molecular map of the region between the distal breakpoints of Df(3R)CA15 and Df(3R)mbc-15A, indicating P1, cosmid, and bacteriophage lambda clones (gray lines). EcoRI sites are indicated. The location of the mbc gene is indicated by a hatched bar, and the direction of transcription is marked by the arrow. (C) A Southern blot of EcoRI/BamHI-digested DNA that was enriched for various deficiency chromosomes, including isogenic ru st e and mbc F6.4 as controls. The test probe is a 2.8-kb EcoRI fragment from P1 clone DS07442 (upper band) while the control probe is a fragment from MHC (lower band).
EMS mutageneses to obtain alleles of mbc have been described (Rushton et al., 1995). In similar screens, ∼11,000 additional chromosomes were analyzed, and 14 new mbc alleles were obtained (Fig. 1). mbc F5.3 was later found to be a small deletion. Additional mutations in this region identify other lethal complementation groups. A subset of these are shown in Fig. 1.
Enriched DNA and Southern Analysis
DNA enriched for the mutant chromosomes was obtained by mating heterozygous (mbc/+) males and females, and collecting the homozygous mutant embryos. Genomic DNA was prepared from unhatched embryos according to Jowett (1986).
Approximately 10 μg of DNA was digested with EcoRI and BamHI, separated on an 0.8% agarose gel, and blotted using the TurboBlotter Rapid Downward Transfer System (Schleicher and Schuell, Inc., Keene, NH). Blots were probed with various genomic fragments from region 95A-C and, as a control for loading, a 2-kb HindIII fragment that includes exons 2, 3a, and 3b from the gene encoding myosin heavy chain (MHC) (Wassenberg et al., 1987). All probes were labeled by random priming (Feinberg and Vogelstein, 1983).
Library Screens, Northern Blots, and DNA Sequencing
Cosmids containing genomic DNA in cytological region 95A-C were obtained from the European Drosophila Genome Project (EDGP). The P1 clone was isolated by the Berkeley Drosophila Genome Project (BDGP) and provided by A. Spradling. Bacteriophage lambda clones were isolated from a Drosophila genomic library in Charon 4 (Maniatis et al., 1978). Fragments were subcloned and analyzed using Southern blots of DNA enriched for the deficiency chromosomes (see above). All DNA between the distal breakpoint of Df(3R)CA15 and the distal breakpoint of Df(3R)mbc-15A was recovered. Genomic fragments containing coding sequence were identified by probing Northern blots (Sambrook et al., 1989).
Subclones of genomic DNA that detected transcribed sequences were used to screen an embryonic 9–12-h cDNA library (Zinn et al., 1988), and several independent cDNA clones were obtained. These included Z5 (nucleotides [nts] 1–2854), Z1.2 (nts 597–1928), Z10b (nts 4184–7040), and Z1.1 and Z6 (nts 6366–7376). DNA fragments from these phages were subcloned into bacterial vectors and sequenced by the Penn State Nucleic Acid Facility. Coding sequence not covered by cDNA clones was obtained from cDNA primed from embryonic RNA and amplified by PCR. In all cases, mbc coding sequence was determined from both DNA strands.
Mutation Detection and Identification
Total RNA was prepared from mbc/isogenic ru st e adults. cDNA was synthesized with Superscript II (GIBCO BRL, Gaithersburg, MD) from various mbc-specific primers. Resulting cDNAs were used to localize mutations within the mbc transcript using the Non-Isotopic RNase Cleavage Assay (NIRCA) of the Mismatch Detect IITM kit (Ambion, Inc., Austin, TX). The mutation in allele mbc F6.4 was detected by a Southern blot of heterozygous mutant DNA digested with EcoRI and BamHI, as described above. The probe was a 6-kb genomic fragment corresponding to the middle of the transcript. Mutant sequences uncovered by either of these analyses were analyzed in DNA enriched for the mbc alleles. Appropriate regions were amplified by PCR and sequenced by the Penn State Nucleic Acid Facility.
Analysis of Maternal and Adult mRNA
Total RNA was prepared from unfertilized eggs, adult males, and adult females (Jowett, 1986). cDNA was synthesized from a primer in the 3′ untranslated region of the mbc gene, and the region between nucleotides 4563 and 5506 was amplified by PCR. Since this region of the mbc transcript spans an intron, contaminating genomic DNA does not give rise to a PCR product. This result was confirmed by amplification of a second region between nucleotides 622 and 1515 (data not shown). To ensure that approximately equal amounts of total RNA were present in each sample, a Northern blot of the original RNA was probed with α1-tubulin (Theurkauf et al., 1986) (data not shown).
Whole Mount Embryo Analysis
Embryos were collected on apple juice/agar plates for 0–6 h and aged as necessary. The embryonic expression pattern of mbc mRNA was determined as described (Tautz and Pfeifle, 1989; Michelson et al., 1990) using a digoxigenin-labeled cDNA fragment (nts 602–1922). mbc-encoded protein was analyzed using a polyclonal rat antiserum (Cocalico Biologicals, Reamstown, PA) that was directed against a 286–amino acid fusion protein. It included amino acids 1717–1970 from the COOH-terminal portion of MBC and was purified from inclusion bodies. Before use, the antiserum was affinity-purified against the original antigen coupled to Affigel 15 (BioRad Labs, Hercules, CA). For confocal studies, anti-MBC was used at a dilution of 1:50, rabbit anti-MEF2 was used at 1:1,000 (Bour et al., 1995), and monoclonal antiphosphotyrosine (Upstate Biotechnology, Inc., Lake Placid, NY) was used at a dilution of 1:100. For detection, fluorescein-conjugated goat anti–rabbit antiserum (Vector Laboratories, Burlingame, CA), fluorescein-conjugated goat anti–mouse antiserum (Rockland Inc., Gilbertsville, PA), and CY-3–conjugated goat anti–rat antiserum (Rockland Inc.) were used, as appropriate. All were preadsorbed overnight on 0–12-h embryos before use. Colorimetric immunohistochemistry used a monoclonal anti-MHC antibody (D. Keihart) at a dilution of 1: 2,000 and an anti–Fasciclin III monoclonal supernatant (Patel et al., 1987) at a dilution of 1:10. These were detected with biotinylated antimouse antiserum and the Vectastain ABC kit (Vector Laboratories). Where necessary, balancer chromosomes were identified by β-galactosidase activity (Klambt et al., 1991) or by colorimetric immunohistochemistry using a mouse monoclonal anti–β-galactosidase antibody (Promega Corp., Madison, WI) at a dilution of 1:1,000. Staining with Texas red–conjugated phalloidin (Molecular Probes, Inc., Eugene, OR) was as described (Ashburner, 1989 b). Four independent experiments were conducted. In two of these studies, wild-type and mutant embryos were treated in parallel throughout the entire analysis but kept in separate tubes. In two subsequent experiments, embryos from a wild-type stock and a balanced mbc mutant stock were pooled. As in the first two experiments, balancer-containing embryos from the mutant stock were identified using anti–β-galactosidase. An average of 30 unstained embryos were mounted and analyzed in each experiment. The results of all four experiments gave comparable results. In the phosphotyrosine experiments, a total of 30 embryos were analyzed in two independent experiments carried out in parallel in separate tubes.
Results
Genetic Localization of mbc
myoblast city was originally identified in a genetic analysis of cytological region 95, on the right arm of the third chromosome (Rushton et al., 1995). Since this is the location of nautilus (nau), the Drosophila homologue of a conserved family of myogenic regulatory genes (Michelson et al., 1990; Paterson et al., 1991), it was of interest to examine genetic lesions in this region for defects in myogenesis. Alleles of mbc were revealed in this analysis since embryos mutant for mbc are characterized by an absence of differentiated muscle fibers and the presence of a correspondingly large number of unfused myoblasts (Rushton et al., 1995). Overlapping deficiencies and EMS-induced point mutations were therefore generated to refine the location of mbc and establish that it represents a novel gene, independent and separate from nau.
The current genetic map of this region is shown in Fig. 1. Df(3R)mbc-30 has been described (Rushton et al., 1995). Df(3R)mbc-15A was generated by γ-irradiation of a homozygous viable P-element insertion in this region. Df(3R)CA15 and Df(3R)CA2 were isolated by imprecise excision of l(3)04684, a homozygous-lethal P-element insertion. Df(3R)CA15 deletes from the P-element insertion toward the distal end of the chromosome. In contrast, Df(3R)CA2 deletes from the P-element toward the centromere. These deficiencies have been examined for the presence or absence of nau sequences by Southern analysis of DNA from embryos homozygous for the deficiencies. By comparison to Df(3R)mbc-30 and Df(3R)mbc-15A, which completely remove nau, neither Df(3R)CA15 nor Df(3R)CA2 appear to remove any known nau sequences (data not shown). Consistent with this genetic map, recent results have established that nau is located in the region centromeric to the proximal breakpoint of Df(3R)CA2 (Keller, C.A., and S.M. Abmayr, unpublished results). Df(3R)mbc-F5.3 actually represents an EMS- induced deletion. It does not complement mbc, as shown in Fig. 1, but contains all known nau sequences (data not shown). Finally, EMS-induced point mutations reveal several additional complementation groups in this region. Other than the transposable element insert TnM2, only those groups distal to the Df(3R)CA2 breakpoint are shown in Fig. 1.
These deficiencies refined the location of mbc to the region between the distal breakpoint of Df(3R)CA15 and the distal breakpoint of Df(3R)mbc-15A and facilitated the cloning of the mbc gene. Of note, several attempts to isolate a P-element insertion in mbc were unsuccessful (data not shown). Therefore, a molecular walk through this region was initiated using DNA fragments isolated from P1 clones and cosmids that have been mapped to region 95BC. Fragments within this genetic interval were identified by Southern analysis of DNA from embryos homozygous for the various deficiency chromosomes. A representative example is shown in Fig. 1 C. The entire region between these deficiency breakpoints is diagrammed in Fig. 1 B and spans ∼34 kb of DNA. As indicated, the organization of these fragments has been confirmed by isolation of bacteriophage lambda clones containing Drosophila genomic DNA.
Identification of the mbc Gene
A single full-length transcript of ∼7.5 kb was detected by Northern analysis throughout development using cloned DNA fragments within the 34-kb region described above. Although full-length clones were not obtained, several small overlapping cDNA clones provided most of the coding sequence of this transcript. The sequence of a small region not covered in the cDNA clones was obtained from embryonic mRNA by reverse transcriptase PCR amplification and sequencing. The embryonic transcript is ∼7.4 kb, with a coding sequence of 5,910 nts. Untranslated regions at the 5′ and 3′ ends of the isolated cDNAs are 560 and 906 bp, respectively. While the genomic organization of mbc has not been analyzed completely, a minimum of eight introns have been identified in a genomic region that spans at least 16 kb. The cDNA sequence has been submitted to GenBank and is not reproduced herein. The deduced amino acid sequence is shown in Fig. 2.
Figure 2.
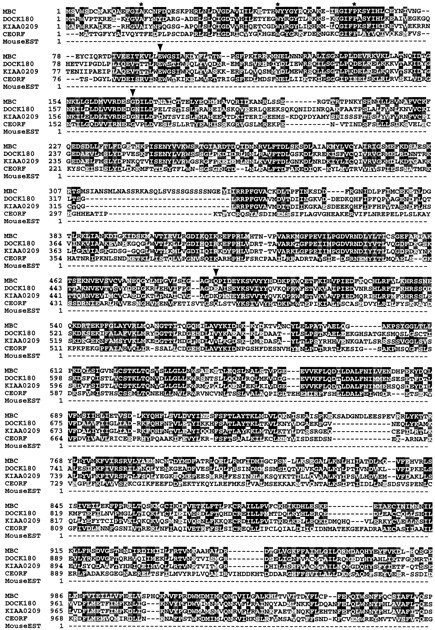

MBC sequence and alignment with human DOCK180 as well as related open reading frames. The alignment was done using Clustal W and presented using Boxshade. Black boxes indicate amino acid identity, while gray boxes indicate amino acid similarity to MBC. Arrowheads highlight mutations found in mbc alleles. The consensus Crk-binding sites (PPxLPxK) of DOCK180 are underlined. A potential Crk-binding site in MBC is noted with dots. Stars mark essential SH3 consensus residues (Musacchio et al., 1994). The GenBank/EMBL/DDBJ accession numbers are D86964 for the myeloblast-specific cDNA KIAA0209, Z81032 and Z81054 for the C. elegans ORFs, and AA110899 for the mouse expressed sequence tag. mbc sequence data are available from GenBank/EMBL/DDBJ under accession number AF007805.
To confirm that this transcript encodes the mbc gene, EMS-induced alleles of mbc were analyzed for sequence alterations. To date, 18 independent alleles of mbc have been generated. Four of these have been described previously (Rushton et al., 1995), while the remaining 14 were generated as part of this analysis. Southern analysis of all alleles was performed to reveal visible rearrangements induced by the chemical mutagen. This analysis uncovered a novel band in an EcoRI/BamHI double digest of DNA from mbc F6.4 (data not shown). Sequence analysis confirmed that the BamHI site had been destroyed by a C to T transition. This missense mutation at amino acid 1579 of the coding sequence changes a proline to a leucine in a conserved region of the protein (Fig. 2, arrowhead).
Additional aberrations were uncovered using a procedure for detecting point mutations that is based on the ability of RNase A to cleave at single base pair mismatches. Several regions of the coding sequence were analyzed by this method and apparent alterations in the candidate sequence were found in three EMS-induced mbc alleles (data not shown). Direct sequencing of these alleles revealed that all were GC to AT transitions at single nucleotides, consistent with the most common form of EMS-induced mutations (Ashburner, 1989 a). These changes resulted in nonsense mutations at amino acid 492 in mbc F12.7 and at amino acid 97 in mbc D11.2. By comparison, mbc I6.6 is a missense mutation at amino acid 168 where a glycine has been replaced by a glutamic acid (Fig. 2).
Structural Homologues of MBC
The size of the mbc-encoded protein is 1,970 amino acids, with a predicted molecular mass of about 226 kD. Data-base homology comparisons using BLAST (Altschul et al., 1990) aligned the MBC protein with DOCK180, a human protein of 1,866 amino acids, with a predicted molecular mass of 215 kD (Hasegawa et al., 1996). DOCK180 was isolated on the basis of an interaction with Crk, a small adapter protein consisting mainly of SH2 and SH3 domains (Reichman et al., 1992; see Discussion). MBC and DOCK180 have significant homology throughout their entire length. In particular, DOCK180 contains a putative SH3 domain that proceeds from amino acids 11–71 and includes the three essential SH3 consensus residues (Musacchio et al., 1994). These three residues, along with several others within this domain, are identical in MBC. DOCK180 contains two copies of the Crk-binding consensus site PPxLPxK (Knudsen et al., 1994; Matsuda et al., 1996), while MBC has one exact and one slightly divergent copy of this consensus site (Fig. 2). By contrast, the putative ATP-binding site noted by Hasegawa et al. (1996) is not conserved. Several additional blocks of homology are present, notably a region in which 24 of 27 amino acids are identical (residues 1566–1592 of MBC). A lesion in the central proline of this block, identified above in mbc F6.4, resulted in a mutant phenotype identical to that previously described (Rushton et al., 1995). This apparent loss-of-function phenotype is identical to that found in alleles with a severely truncated protein (e.g., mbc D11.2; data not shown).
Subsequent BLAST searches also revealed two ORFs with extensive homology to MBC and DOCK180. The first ORF is from a human myeloid cell line, and the second is from the Caenorhabditis elegans genome project. The predicted myeloblast protein is highly homologous to both MBC and DOCK180, while the predicted C. elegans protein is more divergent. Partial sequence from a mouse expressed sequence tag suggests the existence of a murine homologue as well.
Temporal Expression of mbc
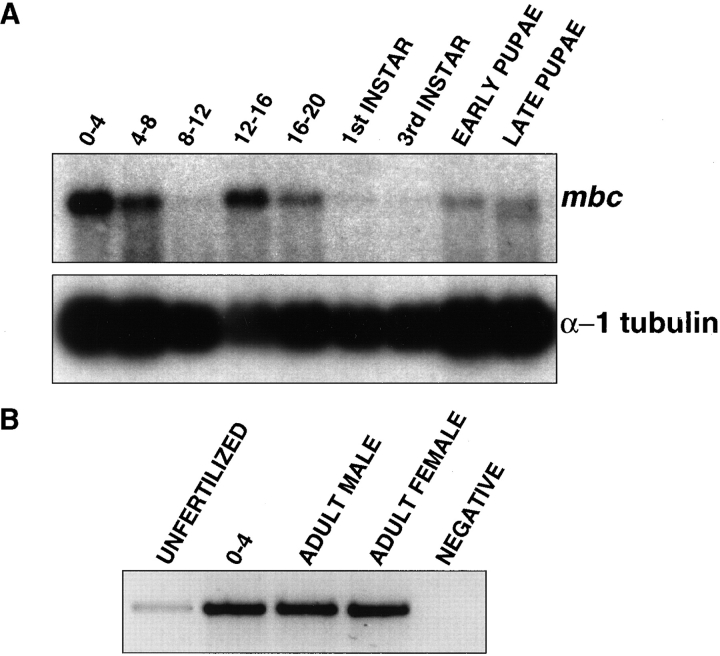
Northern analysis revealed that mbc is expressed early in development, in embryos ∼0–4 h after egg laying. mbc transcript levels remain relatively high during embryogenesis, with the possible exception of a decline from 8–12 h that may be, in part, an artifact of slightly degraded mRNA (Fig. 3 A). Expression was not evident during larval stages, but the transcript does reappear during pupation, suggesting a possible role in adult development. A form of mbc with slightly altered mobility appears late in metamorphosis. This transcript may reflect alternative splicing and is under further investigation (Fig. 3 A, lane 9). PCR amplification of two different regions from the mRNA of unfertilized embryos revealed a small but detectable signal, and suggested that the transcript is maternally provided. Finally, the transcript was expressed in adult males and females, as evidenced by PCR analysis of cDNA (Fig. 3 B).
Figure 3.
Temporal expression of the mbc transcript. (A) A Northern blot containing 4 μg of poly A+ RNA from several stages of development. Probes included a 1.8-kb genomic EcoRI fragment that includes mbc coding sequence and a 400-bp Xba/ HindIII fragment of α1-tubulin (Theurkauf et al., 1986) as a control for loading. (B) Reverse transcriptase PCR analysis of mbc RNA from unfertilized embryos. 0–4-h embryos, adult females, and adult males as detailed in Materials and Methods.
Spatial Expression Pattern of mbc mRNA and Protein during Embryogenesis
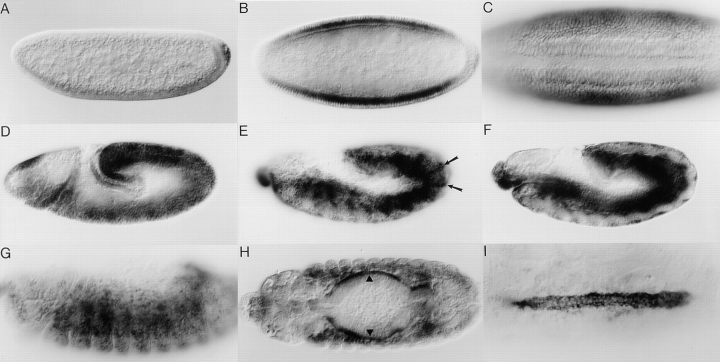
The earliest expression of the mbc transcript is in the pole cells (Fig. 4 A). It is later found in lateral portions of the embryo during cellularization (Fig. 4 B) but is not evident at the termini. Surprisingly, the ventral furrow, which will invaginate during gastrulation to form the mesoderm, shows no expression at this time (Fig. 4 C). At germband elongation, expression is still quite strong in the ectoderm (Fig. 4 D). By late stage 12, the mRNA appears to be decreasing in the ectoderm, leaving a pattern of stripes (Fig. 4 E, arrows). mbc is expressed in both the mesoderm and endoderm during stage 12 (Fig. 4 F). Expression decreases in both the epidermal layer and the somatic mesoderm during stage 14 (Fig. 4 G) but remains strong in the visceral musculature (Fig. 4 H, arrowheads). Examination of a stage 16 embryo revealed mRNA in both the cardial and pericardial cells of the dorsal vessel (Fig. 4 I). Of note, the mbc transcript is not observed in mature muscle fibers.
Figure 4.
Spatial expression pattern of mbc mRNA in wild-type embryos. In all panels, anterior is to the left. In A, D, E, F, and G, dorsal is at the top. (A) Lateral view, early stage 4, before cellularization. (B) Dorsal view, stage 5. (C) Ventral view, stage 6; the invaginating ventral furrow is evident. (D) Lateral view, stage 9. (E) Lateral view of the ectoderm, late stage 12; arrows highlight ectodermal stripes. (F) Lateral view focusing on the mesoderm and endoderm of the same embryo as in E. (G) Lateral view, stage 14; focusing on mesodermal cells. (H) Dorsal view, stage 14; arrowheads indicate the visceral musculature. (I) Dorsal view; stage 16; expression is evident in the cardial and pericardial cells of the dorsal vessel.
The expression pattern of MBC was analyzed by fluorescent immunohistochemistry and confocal microscopy using an antiserum directed against the COOH-terminal portion of the protein. Examination of embryos homozygous for mbc D11.2 confirmed that the antiserum was specific (Fig. 5 A) since this allele encodes a severely truncated form of MBC that would not be detected. While slight temporal differences were evident between maximal levels of mRNA (stage 4; Fig. 4 A) and maximal levels of protein (stage 5; Fig. 5 B) in the pole cells, the expression of the protein essentially correlated with that of the mRNA. MBC appeared to be localized in the cytoplasm (Fig. 5 C), consistent with its human counterpart DOCK180. MBC is also present in the visceral musculature (Fig. 5 D, arrow) and the dorsal vessel (Fig. 5 F, arrow). Cross reactivity of the MBC antiserum was observed in the filtzkorper (Fig. 5 F) but does not correlate with the presence of transcript. Although mRNA was not evident in mature muscles, the protein could be detected at a low level (Fig. 5 E).
Figure 5.
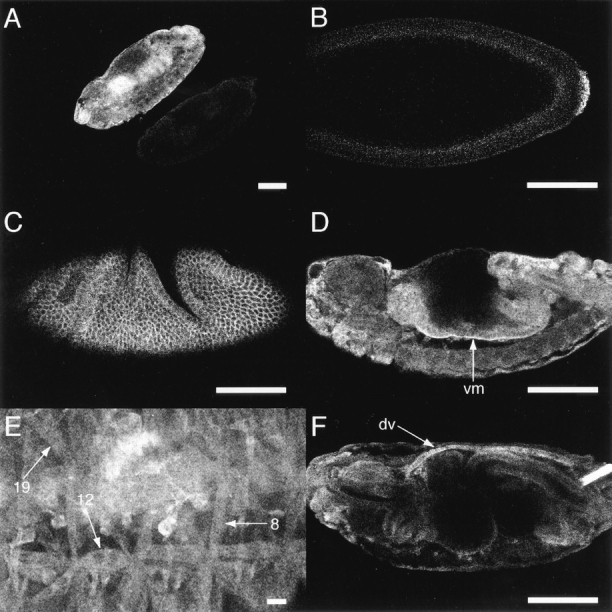
Spatial expression pattern of MBC in wild-type embryos. Anterior is to the left and dorsal to the top in all except A. (A) Stage 13 embryos from the progeny of mbc D11.2/TM3 lacZ-hg stained immunohistochemically for MBC. The embryo to the top left expressed β-galactosidase and therefore carried TM3 lacZ-hg; the embryo to the bottom right (which is barely visible) did not express β-galactosidase (data not shown) and was therefore homozygous for mbc D11.2. As anticipated, no MBC expression is visible in the homozygous mutant embryo, establishing specificity of the antiserum. (B) Wild-type; Lateral view, stage 5. (C) Wild-type; Lateral view, stage 8. (D) Wild-type; Lateral view, stage 14; arrow indicates the visceral musculature (vm). (E and F) Wild-type; Lateral views, stage 16; arrows in E highlight somatic muscles 8, 12, and 19, using the nomenclature of Crossley (1978). Arrow in F marks the dorsal vessel (dv). Bars: (A– D and F) 100 μm; (E) 10 μm.
Fluorescent immunohistochemistry and confocal microscopy were used to confirm that MBC is present in myoblasts. For this analysis, the embryos were hybridized with antibodies to both MBC and MEF2. The mef2 gene encodes a transcription factor that appears to be expressed throughout the mesoderm, including somatic muscle precursors and all muscle fibers (for review see Olson et al., 1995). Nuclei expressing MEF2 were visualized in a late stage 12 embryo in green (Fig. 6 C). By comparison, cytoplasmic expression of MBC was visualized in red (Fig. 6 A). As anticipated from the expression pattern of mRNA, MBC is present in ectodermal and endodermal germ layers. Of note, expression in the ectoderm is concentrated in the epidermal layer and appears to be absent from the underlying neuroectoderm. MBC is also clearly present in presumptive myoblasts, coincident with the MEF2-expressing nuclei (Fig. 6 B).
Figure 6.
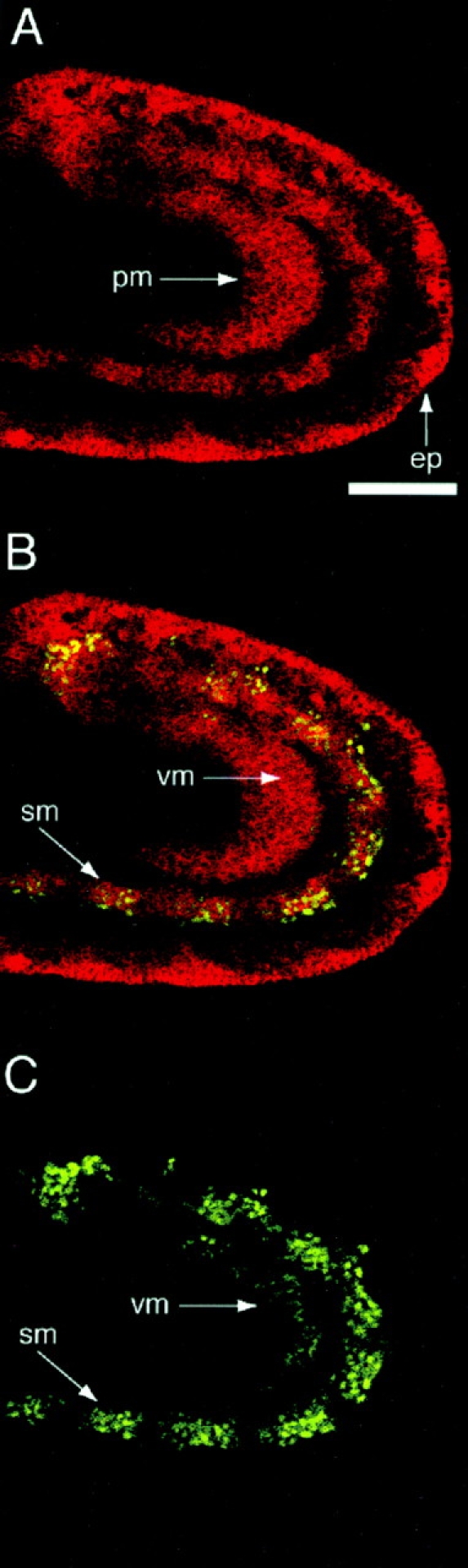
Colocalization of MBC with MEF2 in myoblasts. Using fluorescent immunohistochemistry and confocal microscopy, embryos were analyzed for expression of MBC (red) and MEF-2 (green). The posterior end of an embryo late in stage 12 that is oriented anterior to the left and dorsal to the top is shown. (A) MBC expression. Arrows indicate the posterior midgut primordium (pm) and the epidermis (ep). (B) A composite of A and C, illustrating colocalization in the somatic mesoderm (sm) and the visceral mesoderm (vm). (C) MEF-2 expression. Bar, 50 μm.
Examination of Mesodermal Derivatives in mbc Mutant Embryos
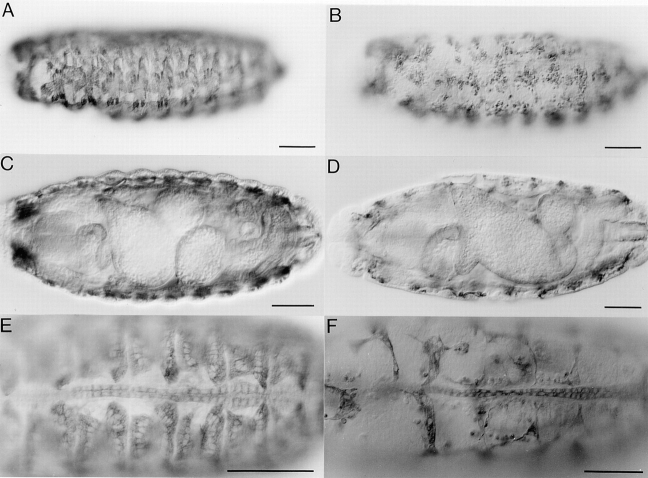
Given the broad expression pattern of mbc, it was of interest to examine mbc mutant embryos for defects in other tissues. For this purpose we used mbc F12.7, since the protein is truncated at amino acid 492, and analyzed embryos that were genetically mbc F12.7/Df(3R)mbc-30. These embryos exhibited the severe somatic muscle phenotype previously reported (Rushton et al., 1995) and shown in Fig. 7 B. By comparison, although the visceral musculature appeared to be present, as evidenced by myosin-staining cells, obvious defects in midgut constriction and orientation were observed in ∼25% of the embryos (Fig. 7, C and D). However, these defects may be an indirect consequence of the lack of somatic muscles rather than a direct effect of the loss of MBC in either the visceral mesoderm or the endoderm. The overall structure of the heart, which expresses MBC late in development, appeared to be normal at this level of analysis (Fig. 7, E and F).
Figure 7.
Analysis of mesodermal derivatives in mbc mutant embryos. Tissues were visualized with a monoclonal antibody to MHC. All embryos are oriented with anterior to the left. A and B are lateral views with dorsal to the top, C and D are ventral views, and E and F are dorsal views. A, C, and E are wild-type embryos; B, D, and F are mbc F12.7/Df(3R)mbc-30 transheterozygotes. (A and B) Somatic muscle pattern of stage 16 embryos. Defects in myoblast fusion, as previously described by Rushton et al. (1995), are evident in B. (C and D) Visceral musculature and gut formation in late stage 16 embryos. Note the midgut constrictions in C and the absence of these constrictions in D. (E and F) Dorsal vessel of stage 17 embryos. At this level, there are no obvious defects. Bars, 50 μm.
Examination of Dorsal Closure and Cytoskeletal Organization in the Epidermis
Although no epidermal defects had been reported in mbc mutant embryos (Rushton et al., 1995), the early expression of mbc in the ectoderm, which persists in the epidermis into stage 14, led us to reexamine mbc mutant embryos for epidermal defects. Visualization of the epidermis with an antibody to Fasciclin III, a glycoprotein on the cell surface (Patel et al., 1987), revealed defects in dorsal closure in ∼80% of the mutant embryos (Fig. 8, E and F). The extent of completion of dorsal closure varied from a relatively small opening surrounded by puckered misshapen cells (data not shown) to a very large opening (Fig. 8 E). In the normal course of dorsal closure in a wild-type embryo, the epidermal cells elongate as shown in Fig. 8 B, and the epithelium stretches over the entire circumference of the embryo (Young et al., 1993). In the early stages of dorsal closure in mbc mutant embryos, the cells along the leading edge of the epidermis appeared to be normal (data not shown). As dorsal closure neared completion, however, many cells along the leading edge ceased to be elongated, adopted a rounded shape, and expressed Fasciclin III abnormally along their migrating edge (Fig. 8, B, D, and F).
Figure 8.
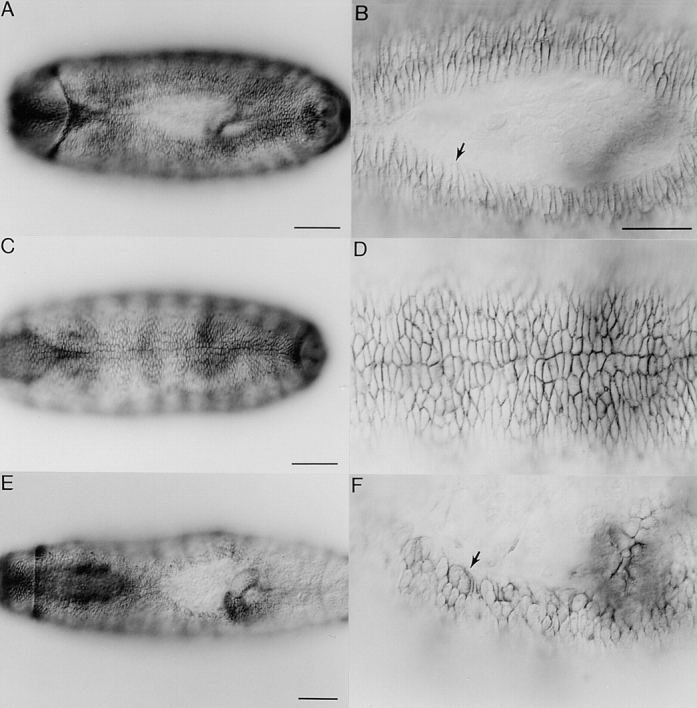
Defects in dorsal closure in mbc mutants revealed by staining with Fasciclin III. All panels are dorsal views with anterior to the left. A–D are wild-type embryos while E and F are mbc F12.7/ Df(3R)mbc-30 transheterozygotes. A and B show a stage 15 embryo in the process of dorsal closure. Arrow in B denotes elongated cells at the leading edge. C and D show a stage 16 embryo that has completed dorsal closure. E and F show a stage 16 embryo that has a pronounced defect in dorsal closure. Arrow denotes cells that are misshapen and have an improper accumulation of Fasciclin III along the leading edge. Bars: (A, C, and E) 50 μm; (B, D, and F) 25 μm.
The cytoskeleton along the leading edge of the epidermis has been implicated in driving the process of dorsal closure (Young et al., 1993). We therefore used fluorescently conjugated phalloidin, which binds filamentous actin, to examine the mbc mutants for defects in cytoskeletal formation and organization. Both wild-type and mbc mutant embryos displayed some variability in the intensity and organization of staining, the range of which is shown in Fig. 9. As shown, the signal in wild-type embryos (Fig. 9 A, a and c) was always stronger than that in mbc mutant embryos (Fig. 9 A, b and d). While frequently more dramatic in cells along the migrating edge, this reduction in signal was also observed throughout the epidermis, consistent with the observed expression of mbc. In addition, it should be noted that ∼20% of the mbc mutant embryos do not exhibit defects in dorsal closure (mentioned above). One might anticipate that these embryos would express relatively normal levels of filamentous actin and exhibit only mild cytoskeletal defects, such as that shown in Fig. 9 A, b. In summary, this analysis suggests that there is a modest but reproducible reduction in cytoskeletal organization in the epidermis of mbc mutant embryos. Unfortunately, examination of the cytoskeletal structure in muscle cells was complicated by the dynamic nature of wild-type muscle cells, making rigorous comparisons with comparable muscle cells in mbc mutant embryos difficult.
Figure 9.
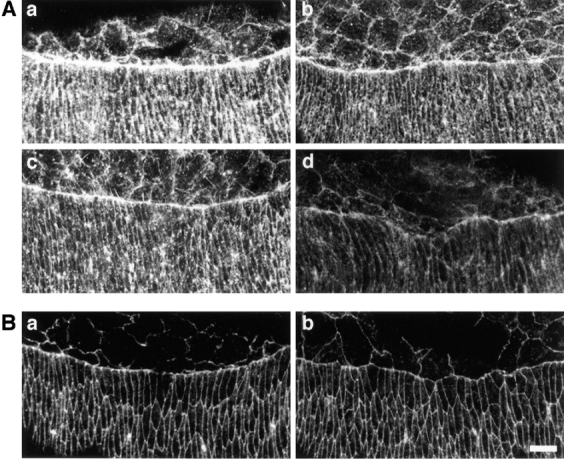
Accumulation of filamentous actin and phosphotyrosine in the epidermis of mbc mutant embryos. Confocal micrographs of embryos stained with Texas red–conjugated phalloidin (A) or an antiphosphotyrosine antibody (B). Anterior is to the left in all panels. A, a and c, and B, a show stage 14 wild-type embryos. A, b and d, and B, b show stage 14 mbc D11.2 homozygous embryos. Bar, 10 μm.
DOCK180, the apparent human homologue of mbc, may be involved in Crk-associated signal transduction from focal adhesions (Hasegawa et al., 1996). If mbc functions in a similar signal transduction pathway in Drosophila, we anticipated that it would be downstream of focal adhesions. Examination of putative focal adhesions in the epidermis of mbc mutant embryos was accomplished using a monoclonal antibody directed against phosphotyrosine, as previously described (Maher et al., 1985; Hanks et al., 1992; Harden et al., 1996). In contrast to the cytoskeletal defects described above, comparison of phosphotyrosine staining patterns in the epidermis of wild-type embryos and homozygous mbc D11.2 embryos revealed no apparent difference during dorsal closure (Fig. 9 B, a and b). This observation is consistent with the possibility that MBC, like DOCK180, is downstream of phosphotyrosine-containing complexes in a signal transduction pathway that, in Drosophila, ultimately affects cell migration, dorsal closure, cytoskeletal organization, and myoblast fusion.
Discussion
The results reported here describe the cloning and characterization of myoblast city, a gene that was initially identified on the basis of a defect in myoblast fusion (Rushton et al., 1995). mbc encodes a novel Drosophila protein with a high degree of homology to the human Crk-associated protein, DOCK180 (Hasegawa et al., 1996). Consistent with that of its human counterpart, mbc expression is not restricted to the somatic mesoderm. Early in development, expression is observed in the pole cells and ectoderm but is absent from the mesodermal epithelium. Later in development, expression is most evident in the epidermis and mesoderm but is absent from neural tissues. The latest detectable expression is in mesodermal derivatives that include the heart and visceral musculature. Consistent with this pattern of expression, defects in myoblast fusion are accompanied by abnormalities in the midgut constrictions and in the ability of the epidermal cells to complete dorsal closure. These cells exhibit alterations in shape, migration, and deposition of Fasciclin III, as well as cytoskeletal organization. Previous studies have reported similar defects for Drac1 (Luo et al., 1994; Harden et al., 1995), the Drosophila homologue of the small GTPase rac1, and imply that mbc may function in the same pathway. Finally, ORFs identified from multiple genome sequencing projects may indicate that MBC and DOCK180 are members of a highly conserved gene family.
The Role of mbc in Ectodermally Derived Tissues
Early expression of mbc in the ectoderm and its persistence in the epidermis led us to examine mbc mutant embryos for epidermal defects. Using Fasciclin III as a marker, we observed that mbc mutant embryos were unable to complete the process of dorsal closure. Contractile filaments formed from actin and myosin are thought to provide the driving force for dorsal closure. Consistent with this suggestion, the absence of nonmuscle myosin in zipper mutant embryos is likely to be responsible for their failure to complete this process (Young et al., 1993). Similarly, overexpression of a form of Drac1 that disrupts both actin and nonmuscle myosin accumulation at the leading edge of the migrating epidermis also inhibits dorsal closure (Harden et al., 1995). Finally, the dorsal closure defects observed in mbc mutants are accompanied by reduced detection of filamentous actin. These results implicate mbc in cytoskeletal organization and dorsal closure and suggest that it may function in the same pathway as Drac1.
Recent studies have shown that Drac1 is necessary for the presence of phosphotyrosine-containing complexes at the leading edge of the epidermis that have been suggested as focal adhesions (Harden et al., 1996). The rho/ rac family of small GTPases has also been implicated in the formation of focal adhesions and in the organization of the actin cytoskeleton in vertebrates (Ridley and Hall, 1992, 1994; Ridley et al., 1992; Nobes and Hall, 1995; Chrzanowska-Wodnicka and Burridge, 1996; for reviews see Clark and Brugge, 1995; Richardson and Parsons, 1995; Takai et al., 1995). The loss of mbc does not appear to affect the formation of these phosphotyrosine-containing complexes, implying that mbc may function downstream of Drac1. This interpretation is consistent with one possible role of human DOCK180 in mediating a signal from focal adhesions to downstream effectors (Hasegawa et al., 1996). Specifically, DOCK180 was isolated on the basis of interaction with the small SH2-SH3 domain-containing adapter protein, Crk (Reichman et al., 1992). Studies addressing the roles of both c-Crk and its oncogenic counterpart v-Crk have suggested an involvement in signal transduction pathways that include receptor tyrosine kinases, ras and MAP kinase, and focal adhesions (Tanaka et al., 1993; Feller et al., 1994; Hempstead et al., 1994; Matsuda et al., 1994; Schaller and Parsons, 1994; Clark and Brugge, 1995; Richardson and Parsons, 1995; Hanks and Polte, 1997). Recently, several proteins have been identified on the basis of interaction with Crk and are likely to be downstream effectors. Among these is the guanine nucleotide exchange factor C3G (Tanaka et al., 1994). Crk may be a critical mediator of signal transduction to events in the nucleus through these molecules. In addition, biochemical evidence has shown that v-Crk and c-Crk can interact with phosphorylated paxillin (Birge et al., 1993; Schaller and Parsons, 1995), one of the components of the focal adhesion (Turner et al., 1990).
It should be noted that although both mbc loss-of-function and dominant negative Drac1N17 embryos exhibit similar defects in dorsal closure and cytoskeletal organization in the epidermis and fusion of myoblasts in the mesoderm (see below), Drac1N17 also induces defects in the peripheral nervous system. In contrast, both the motor neurons and the peripheral nervous system of mbc mutant embryos appear to be normal (Rushton et al., 1995; Prokop et al., 1996; Erickson, M.R.S., and S.M. Abmayr, unpublished observation), consistent with the minimal level of MBC expressed in neural tissues. The simplest interpretation of this apparent inconsistency is that particular factors mediate different aspects of the Drac1 signal transduction cascade. In support of this hypothesis, several different molecules have been identified on the basis of an interaction with vertebrate Crk (Feller et al., 1995, and references therein).
The Role of mbc in Myoblast Fusion
The most apparent mesodermal defect in embryos mutant for the mbc gene is an inability of myoblasts to fuse into muscle fibers, suggesting a role for mbc in the progression of cells from myoblasts to myotubes. This multistep process has been divided into several stages (Knudsen and Horwitz, 1977, 1978; for reviews see Bischoff, 1978; Wakelam, 1985) and includes the acquisition of fusion competence, a time-dependent behavior that may be related to withdrawal from the cell cycle (Bischoff and Holtzer, 1969; Yaffe, 1971; Holtzer et al., 1975b ), myoblast adhesion, and plasma membrane union.
Several features of the mbc-encoded protein seem somewhat inconsistent with a role in either cell adhesion or membrane fusion itself. First, MBC does not have features reminiscent of cell adhesion molecules and appears to be present throughout the cytoplasm rather than membrane bound. Second, both MBC and its structural homologue, DOCK180 (Hasegawa et al., 1996), are expressed in a wide range of tissues that do not fuse. The potential conservation of MBC and DOCK180 in C. elegans, in which the muscle fibers remain mononucleate (Waterston, 1988), is also inconsistent with a direct role for mbc in the fusion process. An alternative possibility is that mbc functions in myoblast differentiation. As mentioned earlier, DOCK180 was identified and subsequently isolated on the basis of interaction with the adapter protein Crk (Reichman et al., 1992). Studies addressing the roles of both c-Crk and v-Crk have implicated these molecules in cell differentiation (Tanaka et al., 1993; Hempstead et al., 1994). Thus, mbc may be essential for a cytoskeleton-related step in differentiation through which, among other things, myoblasts become competent to fuse.
We favor the interpretation that the function of MBC in the mesoderm is analogous to its role in the epidermis and that it functions as an essential intermediate in a signal transduction cascade that also includes the small GTPase Drac1. This pathway could involve tyrosine phosphorylation of complexes that directly modulate events in the cytoskeleton through proteins that include MBC. Alternatively, MBC may function in signal transduction to the nucleus via the ras and MAP kinase pathway and may affect the cytoskeleton only indirectly. Interestingly, while vertebrate studies have not revealed a specific requirement for focal adhesions in myogenesis, they have implicated extracellular matrix components that stimulate focal adhesions, such as fibronectin, in myogenic differentiation (Chen, 1977; Furcht et al., 1978; Menko and Boettiger, 1987; Guan and Shalloway, 1992; Hanks et al., 1992; Enomoto et al., 1993). Additional studies in vertebrates support a role for the cytoskeleton in myoblast fusion. As previously described (see introduction), myoblast fusion is severely limited in the presence of cytochalasin B, an alkaloid that interferes with the assembly of actin filaments (Sanger et al., 1971; Sanger and Holtzer, 1972). While the role of the cytoskeleton in myoblast fusion remains unclear, it may be involved in the formation of lipid-rich domains within the cell membrane that create sites for membrane–membrane fusion. Alternatively, actin filaments may be required for the formation or organization of vesicles that have been observed under the plasma membrane just before fusion of both vertebrate and Drosophila myoblasts (Doberstein et al., 1997; for review see Kalderon, 1980). Interestingly, these vesicles are not observed in mbc mutant embryos, perhaps as a consequence of defects in the actin cytoskeleton (Doberstein et al., 1997).
Additional studies will be necessary to resolve the exact role of mbc in myoblast fusion. In particular, whereas focal adhesions in vertebrates are generally thought to mediate interactions between the cell and the extracellular matrix, no cell–matrix interactions have yet been identified in the Drosophila mesoderm (Tepass and Hartenstein, 1994). In addition, on the basis of examination of phosphotyrosine-containing complexes, our studies seem most consistent with a role for mbc downstream of Drac1 in the epidermis. By comparison, Doberstein et al. (1997) place mbc upstream of a constitutively active form of Drac1. As discussed by these authors, however, the analysis of Drac1 is presently limited to targeted expression of altered forms of the protein and is problematic in the absence of a loss-of-function mutation. It may also reflect a second role for Drac1 in myoblast fusion, not inconsistent with the suggestion that GTPases may act downstream of focal adhesions (Schaller and Parsons, 1994; Clark and Brugge, 1995; Hanks and Polte, 1997). One intriguing possibility consistent with our data and that of Doberstein et al. (1997) is an early requirement for activated Drac1, perhaps to facilitate recruitment of paired vesicles to the membrane via the cytoskeleton, followed by an equally important requirement for Drac1 inactivation later, before fusion. One final issue is that genetic studies have not yet revealed a role for integrin subunits, one of the major components of vertebrate focal adhesions, in myoblast fusion. The larval body wall muscles in embryos mutant for the major integrin subunits, βPS, αPS1, and αPS2, do not appear to exhibit defects in fusion (Brown, 1994; Roote and Zusman, 1995; for reviews see Brown, 1993; Gotwals et al., 1994). However, the number and alternatively spliced forms of integrins identified in Drosophila has continued to increase (Gotwals et al., 1994), and family members that play other roles in myogenesis may yet be isolated. Thus, greater knowledge of GTPases and integrins and the identification of Drosophila homologues to components of vertebrate focal adhesions are likely to refine our working model.
The Role of mbc in Other Tissues
Although mbc is quite highly expressed in the heart and the visceral musculature late in development, these tissues do not appear to be severely affected by the loss of mbc. The visceral musculature does appear to be somewhat defective, as evidenced by the absence of midgut constrictions in a low percentage of embryos, but the heart appears to be relatively normal. One interpretation of such behavior is that another gene, yet to be identified, serves a redundant role in these tissues. Another interpretation is that, while the level of expression observed in unfertilized eggs is quite low, adequate maternally derived MBC protein may be available to embryos lacking zygotic expression of a functional protein. This may be particularly true for the pole cells, which express relatively high levels of MBC early in development.
In summary, we have reported the cloning and characterization of mbc, a novel gene that is essential for events leading to myoblast fusion and dorsal closure. The striking conservation of this molecule with DOCK180, a human gene that may be a target of a signal transduction cascade activated through focal adhesions, suggests the involvement of a signaling cascade in myogenesis, perhaps through organization of the actin cytoskeleton. As a more detailed picture of DOCK180, focal adhesions, and the family of rho/rac like GTPases is revealed, our understanding of the precise role of mbc will grow. Thus, the further identification of common features and homologous genes in different developmental systems may allow us to take advantage of the benefits of each to address the function of a conserved pathway.
Acknowledgments
We thank K. Brubaker, C. Gay, and G. Thomas for assistance with confocal microscopy. We are grateful to D. Heyser and A. Davis for assistance with molecular work, C. Keller and C. Atwell for assistance with deficiency screens, and M. Grill for assistance with point mutations. We thank K. Zinn (California Institute of Technology, Pasadena, CA) for the cDNA library, D. Keihart (Duke University Medical Ctr. Durham, NC) for the MHC antibody, C. Goodman (University of California, Berkeley, CA) for the Fasciclin III antibody, and H. Nguyen (Albert Einstein College of Medicine, Bronx, NY) for the MEF2 antiserum. We thank the Berkeley Drosophila Genome Project for providing P1 clones and P-element insertions and the European Drosophila Genome Project for providing cosmid clones. We are grateful to S. Doberstein (University of California, Berkeley) for information before publication, and C. Keller and B. Bour for critical reading of the manuscript.
Abbreviations used in this paper
- mbc
myoblast city
- MHC
myosin heavy chain
- nau
nautilus
- nts
nucleotides
- ORF
open reading frame
Footnotes
Various portions of this work have been supported by National Science Foundation grant IBN 9204891, National Institutes of Health grant RO1 AR44274, The Muscular Dystrophy Association, and a Junior Faculty Award from the American Cancer Society to S.M. Abmayr.
Please address all correspondence to Susan M. Abmayr, Department of Biochemistry and Molecular Biology and Center for Gene Regulation, The Pennsylvania State University, University Park, PA 16802. Tel.: (814) 863-8254. Fax: (814) 863-7024. E-mail: sma1@psu.edu
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashburner, M. 1989a. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1331 pp.
- Ashburner, M. 1989b. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 434 pp.
- Ball EE, Ho RK, Goodman CS. Muscle development in the grasshopper embryo. I. Muscles, nerves, and apodemes in the metathoracic leg. Dev Biol. 1985;111:383–398. doi: 10.1016/0012-1606(85)90492-0. [DOI] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. . Development (Camb) 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Bate, M. 1993. The mesoderm and its derivatives. In The Development of Drosophila melanogaster. M. Bate and A. Martinez Arias, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1013–1090.
- Birge RB, Fajardo JE, Reichman C, Shoelson SE, Songyang Z, Cantley LC, Hanafusa H. Identification and characterization of a high-affinity interaction between v-Crk and tyrosine-phosphorylated paxillin in CT10-transformed fibroblasts. Mol Cell Biol. 1993;13:4648–4656. doi: 10.1128/mcb.13.8.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, R. 1978. Myoblast fusion. In Membrane Fusion. G. Poste and G.L. Nicolson, editors. North-Holland Publishing Company, New York. 127–179.
- Bischoff R, Holtzer H. Mitosis and the processes of differentiation of myogenic cells in vitro. . J Cell Biol. 1969;41:188–200. doi: 10.1083/jcb.41.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. DrosophilaMEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Brown N. Integrins hold Drosophilatogether. BioEssays. 1993;15:383–390. doi: 10.1002/bies.950150604. [DOI] [PubMed] [Google Scholar]
- Brown NH. Null mutations in the αps2 and βps integrin subunit genes have distinct phenotypes. Development (Camb) 1994;120:1221–1231. doi: 10.1242/dev.120.5.1221. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J.A., and V. Hartenstein. 1985. The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin, Germany. 227 pp.
- Chen LB. Alteration in cell surface LETS protein during myogenesis. Cell. 1977;10:393–400. doi: 10.1016/0092-8674(77)90026-5. [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science (Wash DC) 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- Constantin B, Imbert N, Besse C, Cognard C, Raymond G. Cultured rat skeletal muscle cells treated with cytochalasin exhibit normal dystrophin expression and intracellular free calcium control. Biol Cell. 1995;85:125–135. doi: 10.1016/0248-4900(96)85273-7. [DOI] [PubMed] [Google Scholar]
- Crossley, A.C. 1978. The morphology and development of the Drosophila muscular system. In The Genetics and Biology of Drosophila 2b. M. Ashburner and T.R.F. Wright, editors. Academic Press, London. 499–560.
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuseis required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Auvinen P, Luo L, Jan YN, Simons K. CDC42 and Rac1 control different actin-dependent processes in the Drosophilawing disc epithelium. J Cell Biol. 1995;131:151–164. doi: 10.1083/jcb.131.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson CP, Beckner SK. Activation of myosin synthesis in fusing and mononucleated myoblasts. J Mol Biol. 1975;93:431–447. doi: 10.1016/0022-2836(75)90238-7. [DOI] [PubMed] [Google Scholar]
- Endo T, Nadal-Ginard B. Three types of muscle-specific gene expression in fusion-blocked rat skeletal muscle cells: translational control in EGTA-treated cells. Cell. 1987;49:515–526. doi: 10.1016/0092-8674(87)90454-5. [DOI] [PubMed] [Google Scholar]
- Engel LC, Egar MW, Przybylski RJ. Morphological characterization of actively fusing L6 myoblasts. Eur J Cell Biol. 1985;39:360–365. [PubMed] [Google Scholar]
- Enomoto M, Boettiger D, Menko AS. α5 integrin is a critical component of adhesion plaques in myogenesis. Dev Biol. 1993;155:180–197. doi: 10.1006/dbio.1993.1017. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Feller SM, Ren R, Hanafusa H, Baltimore D. SH2 and SH3 domains as molecular adhesives: the interactions of Crk and Abl. Trends Biochem Sci. 1994;19:453–458. doi: 10.1016/0968-0004(94)90129-5. [DOI] [PubMed] [Google Scholar]
- Feller SM, Knudsen B, Hanafusa H. Cellular proteins binding to the first Src homology 3 (SH3) domain of the proto-oncogene product c-Crk indicate Crk-specific signaling pathways. Oncogene. 1995;10:1465–1473. [PubMed] [Google Scholar]
- Fischman, D.A. 1972. Development of striated muscle. In The Structure and Function of Muscle. G. H. Bourne, editor. Academic Press, New York. 75– 148.
- Fulton AB, Prives J, Farmer SR, Penman S. Developmental reorganization of the skeletal framework and its surface lamina in fusing muscle cells. J Cell Biol. 1981;91:103–112. doi: 10.1083/jcb.91.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furcht LT, Mosher DF, Wendelschafer-Crabb G. Immunocytochemical localization of fibronectin (LETS protein) on the surface of L6 myoblasts: light and electron microscopic studies. Cell. 1978;13:263–271. doi: 10.1016/0092-8674(78)90195-2. [DOI] [PubMed] [Google Scholar]
- Gotwals PJ, Paine-Saunders SE, Stark KA, Hynes RO. Drosophilaintegrins and their ligands. Curr Opin Cell Biol. 1994;6:734–739. doi: 10.1016/0955-0674(94)90101-5. [DOI] [PubMed] [Google Scholar]
- Guan JL, Shalloway D. Regulation of focal adhesion-associated protein tyrosine kinase by both cellular adhesion and oncogenic transformation. Nature (Lond) 1992;358:690–692. doi: 10.1038/358690a0. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Polte TR. Signaling through focal adhesion kinase. BioEssays. 1997;19:137–145. doi: 10.1002/bies.950190208. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Calalb M, Harper MC, Patel SK. Focal adhesion protein-tyrosine kinase phosphorylated in response to cell attachment to fibronectin. Proc Natl Acad Sci USA. 1992;89:8487–8491. doi: 10.1073/pnas.89.18.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden N, Loh HY, Chia W, Lim L. A dominant inhibitory version of the small GTP-binding Rac disrupts cytoskeletal structures and inhibits developmental cell shape changes in Drosophila. . Development (Camb) 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- Harden N, Lee J, Loh HY, Ong YM, Tan I, Leung T, Manser E, Lim L. A Drosophilahomolog of the rac- and cdc42-activated serine/ threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol Cell Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol Cell Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempstead BL, Birge RB, Fajardo JE, Glassman R, Mahadeo D, Kraemer R, Hanafusa H. Expression of the v-crkoncogene product in PC12 cells results in rapid differentiation by both nerve growth factor- and epidermal growth factor-dependent pathways. Mol Cell Biol. 1994;14:1964–1971. doi: 10.1128/mcb.14.3.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzer H, Rubinstein N, Fellini S, Yeoh G, Chi J, Birnbaum J, Okayama M. Lineages, quantal cell cycles, and the generation of cell diversity. Quart Rev Biophys. 1975a;8:523–557. doi: 10.1017/s0033583500001980. [DOI] [PubMed] [Google Scholar]
- Holtzer H, Strahs K, Biehl J, Somlyo AP, Ishikawa H. Thick and thin filaments in postmitotic, mononucleated myoblasts. Science (Wash DC) 1975b;188:943–945. doi: 10.1126/science.1138363. [DOI] [PubMed] [Google Scholar]
- Jowett, T. 1986. Preparation of nucleic acids. In Drosophila: A Practical Approach. D.B. Roberts, editor. IRL Press, Washington, DC. 275–286.
- Kalderon, N. 1980. Muscle cell fusion. In Membrane-Membrane Interactions. N.B. Gilula, editor. Raven Press, New York. 99–118.
- Kalderon N, Gilula NB. Membrane events involved in myoblast fusion. J Cell Biol. 1979;81:411–425. doi: 10.1083/jcb.81.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CA, Erickson MS, Abmayr SM. Misexpression of nautilusinduces myogenesis in cardioblasts and alters the pattern of somatic muscle fibers. Dev Biol. 1997;181:197–212. doi: 10.1006/dbio.1996.8434. [DOI] [PubMed] [Google Scholar]
- Klambt C, Jacobs JR, Goodman C. The midline of the Drosophilacentral nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Feller SM, Hanafusa H. Four proline-rich sequences of the guanine-nucleotide exchange factor C3G bind with unique specificity to the first src homology 3 domain of Crk. J Biol Chem. 1994;269:32781–32787. [PubMed] [Google Scholar]
- Knudsen, K.A. 1991. Fusion of myoblasts. In Membrane Fusion. J. Wilschut and D. Hoekstra, editors. Marcel Dekker, Inc., New York. 601–626.
- Knudsen KA, Horwitz AF. Tandem events in myoblast fusion. Dev Biol. 1977;58:328–338. doi: 10.1016/0012-1606(77)90095-1. [DOI] [PubMed] [Google Scholar]
- Knudsen KA, Horwitz AF. Differential inhibition of myoblast fusion. Dev Biol. 1978;66:294–307. doi: 10.1016/0012-1606(78)90239-7. [DOI] [PubMed] [Google Scholar]
- Lin M-H, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophilatropomyosin gene muscle expression. Proc Natl Acad Sci USA. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: DrosophilaDrac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Maher PA, Pasquale EB, Wang JYJ, Singer SJ. Phosphotyrosine-containing proteins are concentrated in focal adhesions and intercellular junctions in normal cells. Proc Natl Acad Sci USA. 1985;82:6576–6580. doi: 10.1073/pnas.82.19.6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Hardison RC, Lacy E, Lauer J, O'Connell C, Quon D, Sim GK, Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, Nakamura S, Hattori S. Crk protein binds to two guanine nucleotide-releasing proteins for the ras family and modulates nerve growth factor-induced activation of ras in PC12 cells. Mol Cell Biol. 1994;14:5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Ota S, Tanimura R, Nakamura H, Matuoka K, Takenawa T, Nagashima K, Kurata T. Interaction between the amino-terminal SH3 domain of Crk and its natural target proteins. J Biol Chem. 1996;271:14468–14472. doi: 10.1074/jbc.271.24.14468. [DOI] [PubMed] [Google Scholar]
- Menko AS, Boettiger D. Occupation of the extracellular matrix receptor, integrin, is a control point for myogenic differentiation. Cell. 1987;51:51–57. doi: 10.1016/0092-8674(87)90009-2. [DOI] [PubMed] [Google Scholar]
- Michelson AM, Abmayr SM, Bate M, Martinez A, Arias, Maniatis T. Expression of a MyoD family member prefigures muscle pattern in Drosophilaembryos. Genes Dev. 1990;4:2086–2097. doi: 10.1101/gad.4.12a.2086. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Wilmanns M, Saraste M. Structure and function of the SH3 domain. Prog Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of Fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. . Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Paterson BM, Walldorf U, Eldridge J, Dubendorfer A, Frasch M, Gehring WJ. The Drosophilahomologue of vertebrate myogenic-determination genes encodes a transiently expressed nuclear protein marking primary myogenic cells. Proc Natl Acad Sci USA. 1991;88:3782–3786. doi: 10.1073/pnas.88.9.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paululat A, Burchard S, Renkawitz-Pohl R. Fusion from myoblasts to myotubes is dependent on the rolling stone gene (rost) of Drosophila. . Development (Camb) 1995;121:2611–2620. doi: 10.1242/dev.121.8.2611. [DOI] [PubMed] [Google Scholar]
- Prokop A, Landgraf M, Rushton E, Broadie K, Bate M. Presynaptic development at the Drosophilaneuromuscular junction: assembly and localization of presynaptic active zones. Neuron. 1996;17:617–626. doi: 10.1016/s0896-6273(00)80195-6. [DOI] [PubMed] [Google Scholar]
- Reichman CT, Mayer BJ, Keshav S, Hanafusa H. The product of the cellular crkgene consists primarily of SH2 and SH3 regions. Cell Growth Differ. 1992;3:451–460. [PubMed] [Google Scholar]
- Richardson A, Parsons JT. Signal transduction through integrins: a central role for focal adhesion kinase? . BioEssays. 1995;17:229–236. doi: 10.1002/bies.950170309. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. Signal transduction pathways regulating rho-mediated stress fibre formation: requirement for a tyrosine kinase. EMBO (Eur Mol Biol Organ) J. 1994;13:2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Roote CE, Zusman S. Functions for PS integrins in tissue adhesion, migration, and shape changes during early embryonic development in Drosophila. . Dev Biol. 1995;169:322–336. doi: 10.1006/dbio.1995.1147. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophilamuscle development. Development (Camb) 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanger JW, Holtzer H. Cytochalasin B: effects on cell morphology, cell adhesion, and mucupolysaccharide synthesis. Proc Natl Acad Sci USA. 1972;69:253–257. doi: 10.1073/pnas.69.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Holtzer S, Holtzer H. Effects of cytochalasin B on muscle cells in tissue culture. Nature (Lond) 1971;229:121–123. doi: 10.1038/newbio229121a0. [DOI] [PubMed] [Google Scholar]
- Schaller MD, Parsons JT. Focal adhesion kinase and associated proteins. Curr Opin Cell Biol. 1994;6:705–710. doi: 10.1016/0955-0674(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Schaller MD, Parsons JT. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for crk. Mol Cell Biol. 1995;15:2635–2645. doi: 10.1128/mcb.15.5.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Tanaka K, Nakanishi H. Rho as a regulator of the cytoskeleton. Trends Biochem Sci. 1995;20:227–231. doi: 10.1016/s0968-0004(00)89022-2. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Hattori S, Kurata T, Nagashima K, Fukui Y, Nakamura S, Matsuda M. Both the SH2 and SH3 domains of human CRK protein are required for neuronal differentiation of PC12 cells. Mol Cell Biol. 1993;13:4409–4415. doi: 10.1128/mcb.13.7.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, Matsuda M. C3G, a guanine-nucleotide releasing protein expressed ubiquitously, binds to Src homology 3 domains of Crk and Grb2/Ash proteins. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophilaembryos reveals translational control of the segmentation gene hunchback. Chromosoma (Berl) 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tepass U, Hartenstein V. The development of cellular junctions in the Drosophilaembryo. Dev Biol. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Baum H, Bo J, Wensink PC. Tissue-specific and constitutive α-tubulin genes of Drosophila melanogastercode for structurally distinct proteins. Proc Natl Acad Sci USA. 1986;83:8477–8481. doi: 10.1073/pnas.83.22.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CE, Glenney JR, Jr, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelam MJO. The fusion of myoblasts. Biochem J. 1985;228:1–12. doi: 10.1042/bj2280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenberg DR, II, Kronert WA, O'Donnell PT, Bernstein SI. Analysis of the 5′ end of the Drosophilamuscle myosin heavy chain gene. Alternatively spliced transcripts initiate at a single site and intron locations are conserved compared to myosin genes of other organisms. J Biol Chem. 1987;262:10741–10747. [PubMed] [Google Scholar]
- Waterston, R.H. 1988. Muscle. In The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 281–335.
- Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Yaffe D. Developmental changes preceding cell fusion during muscle differentiation in vitro. . Exp Cell Res. 1971;66:33–48. doi: 10.1016/s0014-4827(71)80008-3. [DOI] [PubMed] [Google Scholar]
- Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. A metalloprotease-disintegrin participating in myoblast fusion. Nature (Lond) 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]
- Young PE, Richman AM, Ketchum AS, Kiehart DP. Morphogenesis in Drosophilarequires nonmuscle myosin heavy chain function. Genes Dev. 1993;7:29–41. doi: 10.1101/gad.7.1.29. [DOI] [PubMed] [Google Scholar]
- Zinn K, McAllister L, Goodman CS. Sequence analysis and neuronal expression of fasciclin I in Grasshopper and Drosophila. . Cell. 1988;53:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]