Figure 4.
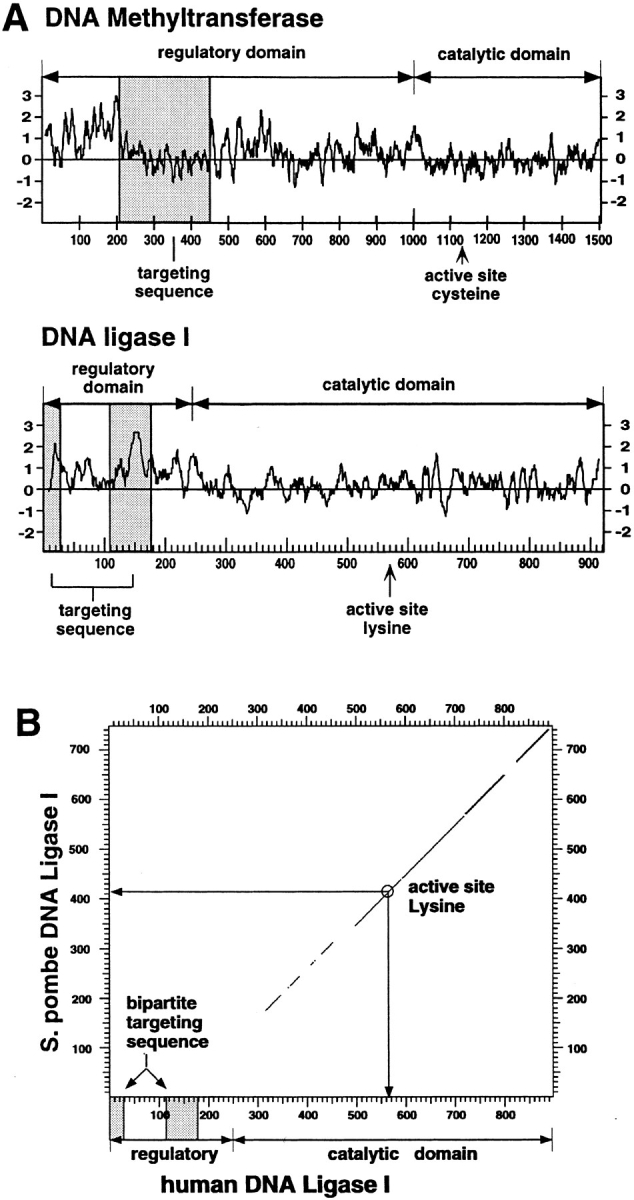
(A) Comparison between the targeting sequences of DNA ligase I and of DNA MTase. A hydrophilicity plot was prepared for both enzymes, and the respective targeting sequences were highlighted by shaded boxes. The overall structure of both enzymes was outlined by delineating the respective regulatory and catalytic domains and indicating the position of the active site residues. The targeting sequences do not share any sequence homology and are on the contrary very different; the bipartite targeting sequence of the DNA ligase I is extremely hydrophilic, while the DNA MTase sequence falls into a rather hydrophobic domain. In both cases the targeting sequence is located in the protease-sensitive, regulatory domain and is dispensable for enzyme activity in vitro. (B) Mammalian DNA ligase-targeting sequence is not conserved in lower eukaryotic homologues. The amino acid sequence of the human DNA ligase I was compared with the Schizosaccharomyces pombe homologue using the DNA Strider program version 1.2 (C. Marck) and the results are displayed in a dot plot format. The yeast and human enzymes show a high degree of homology throughout the catalytic domain, which is outlined in the graph; however, no homologous sequences for the NH2-terminal domain of the human enzyme (including the targeting sequence) could be detected in the fission yeast enzyme. Similar results are obtained comparing the human to the budding yeast protein.
