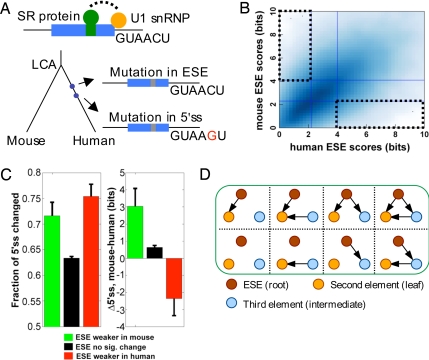
Fig. 1.
The PVSA method for inference of coevolutionary networks. (A) Evolution of a hypothetical exon (blue box) is shown. After the last common ancestor (LCA) of human and mouse, two mutations occur on the human lineage, the first disrupting an ESE, the second strengthening the 5′ss. (B) The PVS for ESE (dotted boxes) is defined as the set of exons whose ESE strength is in the upper 33% of one genome but in the lower 33% of the other genome. (C) Frequency of changes to 5′ss sequences (Left) and changes in 5′ss score (Right) (mean ± SE) in exons belonging to two subsets of the PVS-ESE (green, ESEs weaker in mouse; red, ESEs weaker in human) and to non-PVS-ESE exons (black). (D) Definition of subnetworks in the PVSA method. The subnetworks are rooted directed-acyclic graphs (e.g., with ESE as the root node). For simplicity, only subnetworks with a fixed leaf node (no outgoing edges) are listed.