Abstract
Gain-of-function mutations in the genes encoding the ATP-sensitive potassium (KATP) channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) are a common cause of neonatal diabetes mellitus. Here we investigate the molecular mechanism by which two heterozygous mutations in the second nucleotide-binding domain (NBD2) of SUR1 (R1380L and R1380C) separately cause neonatal diabetes. SUR1 is a channel regulator that modulates the gating of the pore formed by Kir6.2. KATP channel activity is inhibited by ATP binding to Kir6.2 but is stimulated by MgADP binding, or by MgATP binding and hydrolysis, at the NBDs of SUR1. Functional analysis of purified NBD2 showed that each mutation enhances MgATP hydrolysis by purified isolated fusion proteins of maltose-binding protein and NBD2. Inhibition of ATP hydrolysis by MgADP was unaffected by mutation of R1380, but inhibition by beryllium fluoride (which traps the ATPase cycle in the prehydrolytic state) was reduced. MgADP-dependent activation of KATP channel activity was unaffected. These data suggest that the R1380L and R1380C mutations enhance the off-rate of Pi, thereby enhancing the hydrolytic rate. Molecular modeling studies supported this idea. Because mutant channels were inhibited less strongly by MgATP, this would increase KATP currents in pancreatic beta cells, thus reducing insulin secretion and producing diabetes.
Keywords: SUR1, KATP channel, ATP hydrolysis, sulfonylurea receptor
Activating mutations in the ABCC8 and KCNJ11 genes are a common cause of neonatal diabetes (1–3). ABCC8 encodes the regulatory subunit of the ATP-sensitive potassium (KATP) channel. It coassembles with pore-forming Kir6.2 (KCNJ11) subunits to form an octameric channel, with a central tetrameric Kir6.2 pore being surrounded by four SUR1 subunits (4). Both Kir6.2 and SUR1 serve as metabolic sensors, enabling the KATP channel to respond to changes in cellular metabolism. In addition, SUR1 endows the channel with sensitivity to the antidiabetic sulfonylurea drugs.
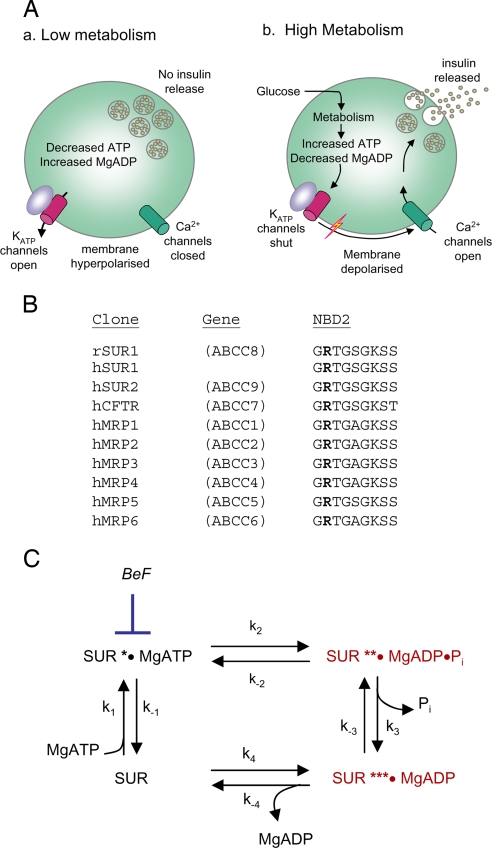
In pancreatic beta cells, metabolic regulation of KATP channels is crucial for glucose-stimulated insulin secretion (Fig. 1A) (3). In unstimulated cells, KATP channels are open and hyperpolarize the membrane enough to keep voltage-gated Ca2+ channels closed. When the plasma glucose concentration rises, glucose uptake and metabolism by the beta cell are enhanced, leading to KATP channel closure. This causes a membrane depolarization that triggers opening of voltage-gated Ca2+ channels and Ca2+-dependent electrical activity. The consequent Ca2+ influx stimulates insulin release.
Fig. 1.
Function and mechanism of the NBDs of SUR1. (A) Role of the KATP channel in glucose-stimulated insulin secretion. (B) Comparison of the amino acid sequences of the Walker A motifs (consensus: GXXXXGKT/S) in the NBDs of various ABCC-family proteins. Residues equivalent to SUR1-R1380 are in boldface. MRP, multidrug resistance-related protein; CFTR, cystic fibrosis transmembrane conductance regulator. (C) The SUR1 ATPase catalytic cycle. BeF, beryllium fluoride (BeF3− and BeF42−).
Metabolic regulation of KATP channel activity is mediated by changes in the concentration of adenine nucleotides, which interact with both Kir6.2 and SUR1 subunits. Binding of ATP to Kir6.2, in a Mg-independent fashion, shuts the channel (5). In contrast, MgADP binding to SUR1 stimulates channel opening (5, 6). Although MgATP can also stimulate KATP channel activity by interaction with SUR1 (7) it must first be hydrolyzed to MgADP (8). KATP channel activity is thus governed by the balance between the inhibitory and stimulatory effects of nucleotides, with low metabolism (low ATP, high MgADP) favoring channel activation and high metabolism (high ATP, low MgADP) favoring inhibition (Fig. 1A).
Here we report a functional mechanism for neonatal diabetes, caused by two different mutations at SUR1 residue 1380, which lies within the second nucleotide-binding domain (NBD2) (9, 10). Our results show that mutation of R1380 to leucine or cysteine results in increased ATPase activity at NBD2. This increase reduces the sensitivity of the channel to inhibition by MgATP and increases the whole-cell KATP current. In pancreatic beta cells, a similar increase in KATP current would be expected to impair insulin secretion and thereby cause diabetes.
Results
Molecular Genetic Analysis.
The R1380C mutation (c.4138C>T) has been reported in three probands with transient neonatal diabetes (9, 10). Two mutations had arisen de novo, but in the other family there was a family history of adult-onset type 2 diabetes or gestational diabetes in mutation carriers. Two pedigrees with the R1380L mutation (c.4139G>T) were identified. The probands and a sibling had diabetes during the neonatal period but inherited the mutation from a parent/grandparent diagnosed with diabetes in adulthood (at 19, 22, 25, and 30 years of age).
Arginine-1380 lies immediately upstream of the Walker A motif of NBD2, which has been implicated in the hydrolysis of MgATP. It is highly conserved and found in the NBD2 of all members of the ABCC subfamily of ATP-binding cassette (ABC) proteins (Fig. 1B), but has not been previously implicated in nucleotide binding or hydrolysis.
ATPase Activity.
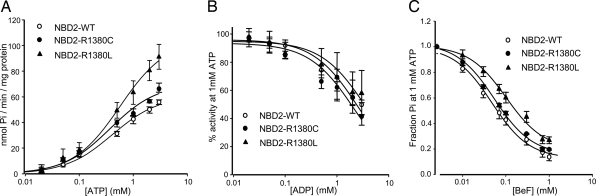
Wild-type NBD2 hydrolyzed ATP with a maximal turnover rate (kcat) of 0.08 s−1 and a Km of 0.41 mM (Fig. 2A and Table 1). The latter is within the range reported previously [0.35–0.6 mM (11, 12)]. Mutation of R1380 to cysteine or leucine did not significantly alter the Km but increased kcat by 1.2-fold (P < 0.05) and 1.7-fold (P < 0.01), respectively (Fig. 2A and Table 1).
Fig. 2.
ATPase activity of SUR1-NBD2 under various conditions. ○, wild-type; ●, R1380C; and ▴, R1380L. (A) ATP activity as a function of ATP concentration. (B) Concentration dependence of MgADP inhibition of ATPase activity (at 1 mM MgATP). (C) Concentration dependence of BeF inhibition of ATPase activity (at 1 mM MgATP).
Table 1.
ATPase activities and kinetic constants
| Construct | Turnover rate,s−1 | Vmax, nmol of Pi·min−1·mg−1 | Km, mM | n |
|---|---|---|---|---|
| NBD2-WT | 0.08 ± 0.002 | 60.8 ± 1.8 | 0.41 ± 0.04 | 4 |
| NBD2-R1380C | 0.09 ± 0.004 | 70.4 ± 3.1* | 0.38 ± 0.05 | 3 |
| NBD2-R1380L | 0.13 ± 0.012 | 104.3 ± 9.9** | 0.55 ± 0.09 | 3 |
WT, wild type. Values are given as mean ± SEM for n separate protein preparations. Significance compared with NBD2-WT: *, P < 0.05; **, P < 0.01.
In all ABC proteins that have been studied, NBD1 and NBD2 associate in a sandwich dimer conformation to form two catalytic ATP-binding sites (site 1 and site 2). These each contain residues from the Walker A and Walker B motifs of one NBD and from the signature sequence of the other NBD (13–16). In our studies, NBD2 will associate as a homodimer. As previously reported (12), mixing wild-type NBD1 and NBD2 did not appear to affect the catalytic activity of either NBD [supporting information (SI) Tables 3 and 4]. Furthermore, both the R1380L and the R1380C mutations increased the ATPase activity of the NBD1–NBD2 mixture (SI Table 3).
Previous studies have shown that MgADP acts as a competitive inhibitor of ATP hydrolysis at NBD2 by trapping the ATPase cycle in the posthydrolytic conformation (Fig. 1C) (8, 12). The nucleotide inhibited ATP hydrolysis by wild-type NBD2 with a Ki of 0.85 mM (Fig. 2B and Table 2). Ki values for the R1380C and R1380L mutants were not significantly different from those of the wild type. There was also no effect of the mutation on MgADP inhibition of ATPase activity when wild-type NBD1 was mixed with wild-type or mutant NBD2 (SI Table 5).
Table 2.
Inhibition constants for ADP and BeF
| Construct | Ki(ADP), mM | Ki(BeF), mM | n |
|---|---|---|---|
| NBD2-WT | 0.85 ± 0.09 | 0.02 ± 0.002 | 4 |
| NBD2-R1380C | 0.68 ± 0.11 | 0.05 ± 0.002* | 3 |
| NBD2-R1380L | 1.36 ± 0.49 | 0.10 ± 0.02** | 3 |
Significance compared with NBD2-WT: *, P < 0.05; **, P < 0.01.
Beryllium fluoride (BeF3− and BeF42−, abbreviated here as BeF) is a potent inhibitor of ATP hydrolysis by many ABC proteins, including the isolated NBD2 of SUR1 and SUR2 (8, 12). It acts by arresting the ATPase cycle in the prehydrolytic conformation (Fig. 1C) (17). The ATPase activity of wild-type NBD2 was potently inhibited by BeF with a Ki of 20 μM, as previously reported (19 μM in ref. 12). Both mutants were significantly less blocked by BeF, the Ki being increased 2.5-fold for R1380C and 5-fold for R1380L (Fig. 2C and Table 2).
KATP Currents.
We next examined the effect of mutating R1380 on KATP currents, by coexpressing wild-type or mutant SUR1 with Kir6.2. We focused on the R1380L mutation, which shows the greatest reduction in ATP hydrolysis.
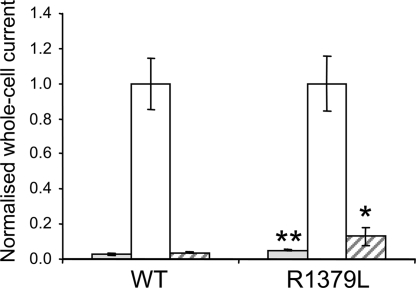
Fig. 3 shows that whole-cell KATP currents are very small under resting conditions, presumably because of the high intracellular ATP concentration, but are dramatically increased by sodium azide, which inhibits mitochondrial metabolism. Resting R1380L currents were slightly (2-fold), but significantly (P < 0.01), larger than wild type. They were further increased by metabolic inhibition, indicating that the channel is only partially closed at resting ATP. The sulfonylurea tolbutamide blocked wild-type currents by 96 ± 1% (n = 12) and R1380L currents by 87 ± 5% (n = 13) (P < 0.05) (Fig. 3). This finding suggests that the diabetes of patients carrying these mutations should be treatable with sulfonylureas.
Fig. 3.
Mean steady-state whole-cell KATP currents evoked by a voltage step from −10 to −30 mV before (control, gray bars) and after (white bars) application of 3 mM sodium azide, and in the presence of 3 mM azide plus 0.5 mM tolbutamide (hatched bars) for wild-type channels (n = 12) and R1380L channels (n = 13). *, P = 0.05; **, P = 0.01 compared with control (t test).
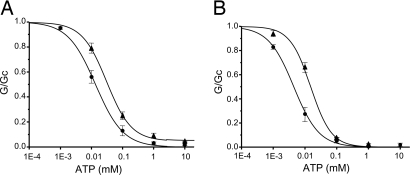
As Fig. 4A shows, R1380L channels were less ATP sensitive than wild type, when measured in inside-out patches. The concentration of ATP causing half-maximal block (IC50) increased from 16 μM to 35 μM when R1380 was mutated (SI Table 6). Furthermore, the amount of current that remained unblocked at physiological MgATP concentrations (1–10 mM) increased from <1% of maximal for wild-type channels to 5% for R1380L channels at 3 mM MgATP.
Fig. 4.
Mean relationship between ATP concentration and KATP conductance (G), expressed relative to that in the absence of nucleotide (Gc) for wild-type (•) and R1380L (▴) channels. (A) In the presence of Mg2+: wild-type channels (n = 10) and R1380L channels (n = 5). (B) In the absence of Mg2+: wild-type channels (n = 7) and R1380L channels (n = 7).
Although ATP is thought to influence KATP channel activity in Mg2+-free solutions only via Kir6.2, the ATP sensitivity of the mutant channel in the absence of Mg2+ also differed from that of wild type (Fig. 4B). The IC50 increased ≈3-fold, from 5 μM to 16 μM (Fig. 4B, SI Table 6). The intrinsic open probability (Po) (i.e., that measured in the absence of nucleotide) was unaffected by the mutation, being 0.21 ± 0.03 (n = 8) for wild-type compared with 0.28 ± 0.06 (n = 6) for R1380L channels. These results contrast with some other SUR1 ND mutations, which reduce the ATP sensitivity of the KATP channel in Mg2+-free solutions by impairing gating (18).
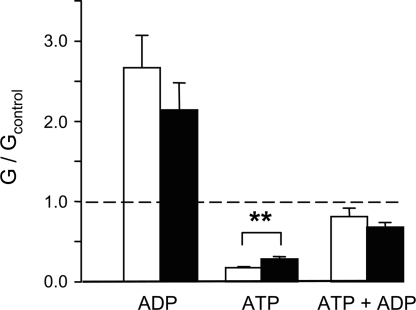
Finally, no significant difference was observed in the extent of channel activation by MgADP in either the presence or absence of ATP (Fig. 5), consistent with the fact that the Ki for MgADP inhibition of ATP hydrolysis remained unchanged (Fig. 2B).
Fig. 5.
Mean KATP currents recorded in the presence of 0.1 mM ADP, 0.1 mM ATP, or 0.1 mM ATP plus 0.1 mM ADP. Currents (G) are expressed relative to the mean of the currents recorded in control solution (Gc) before and after application of nucleotide. **, P < 0.01.
Structural Considerations.
The three-dimensional structure of SUR1 at atomic resolution is unknown. However, crystal structures of the NBDs of many ABC proteins have been solved (14–16). These all share the same overall fold, suggesting that homology models based on these structures may provide a reasonable approximation to the backbone structure of SUR1.
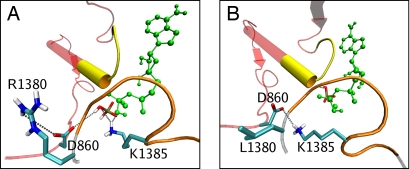
A homology model of the NBD heterodimer of SUR1 based on the crystal structure of Sav1866 (15, 16) (32% sequence identity, see SI Text) shows that R1380 lies within NBD2, ≈10 Å away from the phosphate tail of ATP (Fig. 6A). However, neither its side chain nor its backbone contacts the nucleotide. Instead, R1380 appears to interact with a conserved aspartate (D860) in NBD1, which interacts in turn with the γ-phosphate of ATP (Fig. 6A) (13). An aspartate is also present at an equivalent position in the NBD2 homodimer (D1512).
Fig. 6.
Wild-type and mutant ATP-binding site 2. (A) Homology model of ATP-binding site 2 of wild-type SUR1, indicating the main interactions of the posthydrolytic inorganic phosphate (CPK colors). MgADP is green, the NBD1 backbone is red, the signature sequence of NBD1 is yellow, the NBD2 backbone is gray, and the Walker A domain of NBD2 is orange. (B) Mutant model, with the posthydrolytic phosphate unbound. Unlike the wild-type simulation, K1385 hydrogen bonds to D860.
Molecular dynamics simulations were carried out on wild-type and mutant models. To model the posthydrolytic state, MgADP was placed in its binding site and an inorganic phosphate (Pi) was placed where the γ-phosphate of MgATP would be located (see SI Text). The simulations show a clear difference in the interaction of Pi in wild-type and mutant models. In the wild-type model, Pi interacts stably with surrounding residues, forming hydrogen bonds with K1385 (during 70% of a 10-ns simulation) and D860 (20%) (Fig. 6A). R1380 seems to coordinate D860 through hydrogen bonding of the side chains. In contrast, in the R1380L mutant simulation, Pi quickly loses its interaction with these residues and becomes almost completely hydrated within 6 ns. It forms a weak association with Q1427, but whether this interaction is biologically relevant is less clear (Fig. 6B). A more detailed analysis shows that the R1380L mutation does not appear to affect MgADP binding (see SI Text), although the simulations are too short to be certain. Overall, the molecular dynamics simulations support the idea that the off-rate for Pi is enhanced by the R1380L mutation. This appears to result from loss of the R1380–D860 interaction, which probably affects association of D860 and Pi.
Discussion
We have identified a mechanism by which mutations in SUR1 give rise to neonatal diabetes: enhanced ATPase activity at NBD2. To our knowledge, this is the first report of a mutation that enhances the ATPase activity of an ABC protein.
ATPase Activity.
Mutation of R1380 to C or L increased the ATPase activity of NBD2 of SUR1, as measured by increased Pi production. Both the Km for MgATP and the Ki for MgADP inhibition were largely unchanged. The fact that MgADP activation was similar for both mutant and wild-type KATP channels supports the idea that MgADP interaction with NBD2 is unaffected. These data therefore suggest that the observed increase in turnover rate produced by the R1380C and R1380L mutations is primarily a result of an increased hydrolysis of bound MgATP and/or an increased off-rate of Pi (k3 in Fig. 1C). Both mutants bound BeF with lower affinity than wild type. BeF, in combination with MgADP, mimics the γ-phosphate in ATP before hydrolysis and thus traps the cycle in a pseudo-prehydrolytic state. A lower affinity for BeF would be consistent with a lower affinity for Pi and a faster off-rate.
A caveat with these studies is that ATPase activity was measured on isolated NBD2. However, the wild-type data are in good agreement with those we obtained in electrophysiological studies using full-length SUR1. Moreover, mixing NBD2 (wild-type or mutant) with NBD1, to facilitate formation of NBD1–NBD2 heterodimers, gave results qualitatively similar to those from the wild-type or mutant NBD2 homodimers.
In both our model and the crystal structure of the bacterial ABC protein Sav1866 (16), Arg-1380 is ≈10 Å away from the γ-phosphate of ATP. It is therefore unlikely to interact with Pi directly. Our molecular dynamics simulations suggest that the mutation in R1380 disturbs γ-phosphate binding by the loss of the R1380–D860 interaction, which affects D860 association with the phosphate. This possibility is consistent with the functional data.
How does the enhanced rate of ATP hydrolysis at NBD2 favor channel activation? It is now well established that channel activation is facilitated by the presence of MgADP at NBD2, and thus we predict that both mutations will enhance the amount of time spent in the MgADP-bound state. Our results demonstrate that MgADP binding (and transduction) are unaffected, but that ATP hydrolysis (Pi release) is increased. The simplest way to account for these data is to speculate that speeding up the ATPase cycle ensures that more time is spent in the MgADP-bound state and less in the unoccupied (apo) state or MgATP-bound (prehydrolytic state).
In most ABC proteins, R1380 is highly conserved. It has not been implicated previously in binding or catalysis of ATP. Interestingly, when the equivalent residue in SUR2B (R1344) is mutated to alanine, MgADP is no longer able to enhance channel activity preblocked by MgATP (19). Thus the behavior of SUR2B-R1344L channels resembles that of wild-type SUR2A channels. It has therefore been postulated that R1344 may interact with the last 42 amino acids of SUR2, thereby influencing the conformation of the Walker A loop and its functional properties. The fact that the C terminus of SUR1 has a different sequence from both SUR2A and SUR2B may explain why the SUR1-R1380 mutation does not alter MgADP activation.
Channel Activity.
The extent of inhibition by MgATP reflects the balance between the inhibitory effect of ATP (via Kir6.2) and the stimulatory effect of MgATP (via SUR1). Any increase in MgATP activation would therefore bias the MgATP concentration–inhibition curve toward higher ATP concentrations. Thus the lower MgATP sensitivity of R1380L channels is consistent with the enhanced rate of MgATP hydrolysis we measure and our suggestion that the dwell time in the MgADP-bound state of the ATPase cycle is increased.
Nucleotide activation of KATP currents requires Mg2+. Thus if the sole effect of the R1380L mutation were to enhance MgATP hydrolysis at NBD2 of SUR1, there should be no difference in the ATP sensitivity of wild-type and R1380L channels in the absence of Mg2+. Unexpectedly, however, mutant channels showed a lower sensitivity to ATP in the absence of Mg2+. This is not a consequence of an increase in Po (20), which was unchanged. Therefore the mutation may affect channel activation either by modification of the ATP-binding site on Kir6.2 or by binding of ATP in a Mg-independent fashion to NBD1.
R1380L/C and Neonatal Diabetes.
Our data show that mutations at R1380 have modest (<2-fold) effects on the rate of ATP hydrolysis, the ability of ATP to block the KATP channel, and the resting whole-cell KATP current. However, because the KATP channel dominates the membrane potential of the beta cell, even a small increase in KATP current can cause a hyperpolarization sufficient to inhibit electrical activity and insulin secretion (21). Previous studies of Kir6.2 mutations have shown a good correlation between the magnitude of the KATP current at physiological levels of MgATP and the severity of the clinical phenotype. It is still too early, however, to say whether such a correlation holds for SUR1 mutations.
Interestingly, some patients with mutations at codon 1380 have transient neonatal diabetes but their relatives have permanent diabetes diagnosed in adulthood (10). This pattern of transient neonatal diabetes in a proband and permanent diabetes diagnosed in childhood or adulthood has been reported in 12 families with 10 different mutations in the KCNJ11 or ABCC8 genes (9, 10). Birth weight, a reflection of insulin-mediated growth and thus insulin secretion in utero, is reduced to a similar extent whether the patient is diagnosed before 6 months or not (10). These data, together with the similar phenotypes observed between the probands, suggests that genetic modifiers are unlikely to play a large role in phenotypic variability. Rather, our findings imply that KATP channel mutations have a biphasic course, and that patients diagnosed later in life may have had a period of hyperglycemia that was undetected in the neonatal period.
The in vitro response of mutant channels to tolbutamide predicts that patients with an R1380C or R1380L mutation will respond to sulfonylureas. This is indeed the case, because of the nine family members currently studied, five are now treated with sulfonylureas, whereas four have yet to attempt transfer off insulin (9, 10).
Conclusions.
In this study, we have examined the link between mutations in SUR1 and neonatal diabetes, with reference to mutations in the Walker A arginine-1380 of NBD2. We have shown that an increase in KATP current can be attributed, in part, to an increase in the ATP hydrolysis activity of the nucleotide-binding domains of SUR1.
Materials and Methods
Molecular Genetic Analysis.
Genomic DNA was extracted from peripheral leukocytes by using standard procedures. The ABCC8 gene was amplified and sequenced in probands as described in ref. 10. Results were compared with the published sequence NM_000352.2 with the alternatively spliced residue in exon 17 (L78208, L78224). Compared with the reference sequence used by Babenko et al. (9), this includes an additional amino acid. Hence the cysteine mutation at residue 1380 described in this article has previously been reported by Babenko et al. (9) as affecting R1379.
NBD2 Mutation and Expression.
The C terminus of maltose-binding protein was fused to the N terminus of the second nucleotide-binding domain of rat SUR1 (residues Lys-1319 to Lys-1581) by using the pMal-c2 vector system (New England Biolabs) and expressed as a fusion protein, abbreviated here as NBD2. Site-directed mutagenesis was carried out according to the manufacturer's instructions (QuikChange SDM kit; Stratagene) and confirmed by sequencing. Plasmids were transformed into BL21-CodonPlus E. coli cells (Stratagene), and wild-type and mutant proteins were purified as described previously (12). Typical yields were 1.0 mg of protein per liter.
Nucleotide Hydrolysis.
ATPase activity was measured with a colorimetric assay for liberated Pi, as described in ref. 22. All assays were performed at 37°C. Details of control experiments are described in ref. 22 and SI Figs. 7 and 8.
Experimental repeats (n) refer to separate protein preparations. All protein preparations were assayed in duplicate. Values are given as mean ± SEM. The Michaelis–Menten equation was fit to concentration–activity relationships and used to obtain the Km. Turnover rate (nmol of Pi released per second per nmol of protein) was calculated using molecular masses of 87 kDa for MBP-NBD1 and 74 kDa for MBP-NBD2. The IC50 for MgADP and BeF inhibition were calculated by fitting the data to the Langmuir equation: y = B + {1/[1 + ([I]/IC50)]}, where y is the ATP hydrolysis rate, IC50 is the concentration of inhibitor, I, at half-maximal inhibition, and B is the level of remaining ATPase activity at maximal inhibition (where B = 0 for complete inhibition). Ki values were then calculated from the IC50 by using the equation for competitive inhibition of Cheng and Prusoff (23): Ki = IC50/(1 + {[ATP]/Km(ATP)}).
Electrophysiology.
Wild-type or mutant rat SUR1 was coexpressed with human Kir6.2 in Xenopus oocytes. Mutant channels (which contain four mutant SUR1 subunits and four wild-type Kir6.2 subunits) are referred to in this paper as R1380L or R1380C channels. Whole-cell currents were recorded by two-electrode voltage clamp (20). Whole-cell currents were normalized to their value in the presence of azide, to take account of difference in expression between different batches of oocytes.
Macroscopic currents were recorded from inside-out membrane patches (20). The pipette solution contained 140 mM KCl, 1.2 mM MgCl2, 2.6 mM CaCl2, and 10 mM Hepes (pH 7.4 with KOH). The intracellular (bath) solution contained 107 mM KCl, 1 mM K2SO4, 2 mM MgCl2, 10 mM EGTA, 10 mM Hepes (pH 7.2 with KOH), and MgATP as indicated. ATP concentration–response curves were fit with the Hill equation:
where [ATP] is the ATP concentration, IC50 is the ATP concentration at which inhibition is half maximal, h is the slope factor, a represents the fraction of unblocked current at saturating [ATP], and G is the KATP conductance. To control for rundown, Gc was taken as the mean of the conductance in control solution before and after ATP application. Single-channel currents were recorded at −60 mV from inside-out patches, as described in ref. 20. All data are given as mean ± SEM. Error bars are shown where they are larger than the symbols. Significance was evaluated by using Student's t test.
Molecular Modeling.
A molecular model of the SUR1 NBDs was constructed based on the crystal structure of Sav1866 (16), and molecular dynamics simulations of 9- to 16-ns duration were carried out by using the GROMACS software (24). Full details of the modeling are given in the SI Text.
Supplementary Material
Acknowledgments
This research was supported by the Wellcome Trust (F.M.A., M.S.P.S., AT.H., and A.-M.P.) and the European Union [F.M.A. and A.T.H., EuroDia-(LSHM-CT-2006-518153)]. F.M.A. is a Royal Society Research Professor. H.d.W. is a Wellcome Trust Training Fellow and is supported by the Wellcome Trust OXION Initiative in Ion Channels and Disease. J.A. is a Wellcome Trust Structural Biology Research Student. A.T.H. is a Wellcome Trust Clinical Research Leave Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707428104/DC1.
References
- 1.Ashcroft FM. J Clin Invest. 2005;115:2047–2058. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, et al. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 3.Hattersley AT, Ashcroft FM. Diabetes. 2005;54:2503–2513. doi: 10.2337/diabetes.54.9.2503. [DOI] [PubMed] [Google Scholar]
- 4.Mikhailov MV, Campbell JD, de Wet H, Shimomura K, Zadek B, Collins RF, Sansom MSP, Ford RC, Ashcroft FM. EMBO J. 2005;24:4166–4175. doi: 10.1038/sj.emboj.7600877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker SJ, Gribble FM, Zhao C, Trapp S, Ashcroft FM. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 6.Nichols CG, Shyng SL, Nestorowicz A, Glaser B, Clement JP, Gonzales G, Aguilar-Bryan L, Permutt MA, Bryan J. Science. 1996;272:1785–1787. doi: 10.1126/science.272.5269.1785. [DOI] [PubMed] [Google Scholar]
- 7.Gribble FM, Tucker SJ, Haug T, Ashcroft FM. Proc Natl Acad Sci USA. 1998;95:7185–7190. doi: 10.1073/pnas.95.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zingman LV, Alekseev AE, Bienengraeber M, Hodgson D, Karger AB, Dzeja PP, Terzic A. Neuron. 2001;31:233–245. doi: 10.1016/s0896-6273(01)00356-7. [DOI] [PubMed] [Google Scholar]
- 9.Babenko AP, Polak M, Cave H, Busiah K, Czernichow P, Scharfmann R, Bryan J, Aguilar-Bryan L, Vaxillaire M, Froguel P. N Engl J Med. 2006;355:456–466. doi: 10.1056/NEJMoa055068. [DOI] [PubMed] [Google Scholar]
- 10.Flanagan SE, Patch A-M, Mackay DJ, Edghill EL, Gloyn AL, Robinson D, Shield JP, Temple K, Ellard S, Hattersley AT. Diabetes. 2007;56:1930–1937. doi: 10.2337/db07-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masia R, Enkvetchakul D, Nichols CG. J Mol Cell Cardiol. 2005;39:491–501. doi: 10.1016/j.yjmcc.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 12.de Wet H, Mikhailov MV, Fotinou C, Dreger M, Craig TJ, Vénien-Bryan C, Ashcroft FM. FEBS J. 2007;274:3532–3544. doi: 10.1111/j.1742-4658.2007.05879.x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell JD, Proks P, Lippiat JD, Sansom MS, Ashcroft FM. Diabetes. 2004;53:S123–S127. doi: 10.2337/diabetes.53.suppl_3.s123. [DOI] [PubMed] [Google Scholar]
- 14.Locher KP. Curr Opin Struct Biol. 2004;14:426–431. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Dawson RJ, Locher KP. Nature. 2006;443:156–157. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 16.Dawson RJ, Locher KP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 17.Werber MM, Peyser YM, Muhlrad A. Biochemistry. 1992;31:7190–7197. doi: 10.1021/bi00146a023. [DOI] [PubMed] [Google Scholar]
- 18.Proks P, Shimomura K, Craig TJ, de Wet H, Girard CAJ, Ashcroft FM. Hum Mol Genet. 2007;16:2011–2019. doi: 10.1093/hmg/ddm149. [DOI] [PubMed] [Google Scholar]
- 19.Matsushita K, Kinoshita K, Matsuoka T, Fujita A, Fujikado T, Tano Y, Nakamura H, Kurachi Y. Circ Res. 2002;90:554–561. doi: 10.1161/01.res.0000012666.42782.30. [DOI] [PubMed] [Google Scholar]
- 20.Proks P, Antcliff JF, Lippiat J, Gloyn AL, Hattersley AT, Ashcroft FM. Proc Natl Acad Sci USA. 2004;101:17539–17544. doi: 10.1073/pnas.0404756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarasov AI, Welters HJ, Senkel S, Ryffel GU, Hattersley AT, Morgan NG, Ashcroft FM. Diabetes. 2006;55:3075–3082. doi: 10.2337/db06-0637. [DOI] [PubMed] [Google Scholar]
- 22.Chifflet S, Torriglia A, Chiesa R, Tolosa S. Anal Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Y, Prusoff WH. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 24.Berendsen HJC, van der Spoel D, van Drunen R. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.