Figure 1.
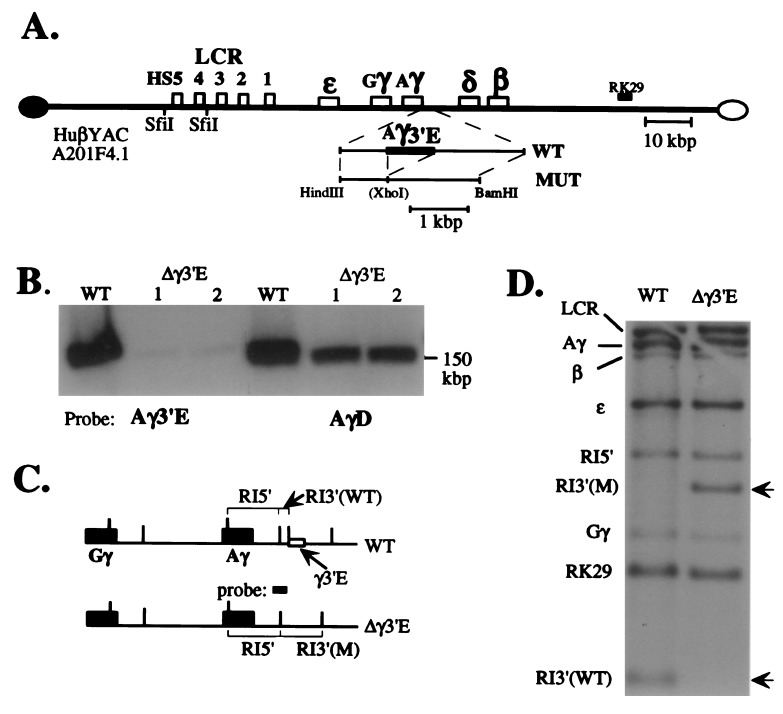
Generation and characterization of the 3′ γ-globin element deletion mutant YAC. (A) Diagrammatic representation of wild-type and mutant YACs. An expanded diagram depicting the targeting construct generated here (abbreviated MUT) is shown below the diagram of the wild-type YAC (the position of the Aγ-3′ element deletion is indicated by a fusion within the MUT subclone). The position of the probes used in Fig. 2 (below) are indicated. (B) Wild-type and two separate subclones of the mutant YAC DNAs (1 and 2) were separated from yeast chromosomes by PFGE, transferred to nylon filters, and hybridized to either the Aγ 3′ element (Aγ3′E) or to a sequence lying 3′ to the Aγ3′E (called “AγD”; nucleotides 44,350–44,750 of the locus; www.globin.cse.psu.edu) to verify that the Aγ3′E was deleted specifically and that the gross size of the mutant YAC was not altered during the manipulations (PFGE does not resolve the difference in size between the two 150-kbp YACs ± the 757-bp Aγ3′E). Faint hybridization is detected in the Aγ3′E deletion mutant lanes with the Aγ3′E probe due to homology with the corresponding Gγ sequence (27), which shows no biological activity (4). (C) A diagram depicting both the wild-type and predicted mutant YAC structures in the vicinity of the γ-globin genes. Recognition sites for EcoRI are depicted as vertical lines. The “probe” (spanning the 3′ gene-proximal EcoRI site) referred to here and in D was generated from the genomic locus from nucleotides 40,861–41,867. (D) A Southern blot showing that the wild-type and Aγ3′E mutant YACs bear identical EcoRI fragments hybridizing to the LCR (10.4 kbp), RK29 (1.0 kbp), ɛ- (3.7 kbp), γ- (6.9 kbp), and β-globin (5.5 kbp) genes (see Fig. 2A). Two different EcoRI fragments hybridize to a probe spanning part of the deletion (C). Note that the Aγ3′E deletion results in the loss of a 0.4-kbp EcoRI fragment, simultaneously creating a 2.1-kbp EcoRI fragment. This probe (C) also crosshybridizes with a Gγ-globin 3′ fragment, 1.6 kbp in size.