Abstract
Vesicomyid clams depend entirely on sulfur-oxidizing endosymbiotic bacteria for their nutriment. Endosymbionts that are transmitted cytoplasmically through eggs, such as these, should exhibit a phylogenetic pattern that closely parallels the phylogeny of host mitochondrial genes. Such parallel patterns are rarely observed, however, because they are obscured easily by small amounts of horizontal symbiont transmission or occasional host switching. The present symbiont genealogy, based on bacterial small subunit (16S) rDNA sequences, was closely congruent with the host genealogy, based on clam mitochondrial cytochrome oxidase subunit I and large subunit (16S) rDNA sequences. This phylogenetic evidence supports the hypothesis of cospeciation and a long term association between the participants in this symbiosis.
The discovery of dense communities of animals associated with deep sea hydrothermal vents profoundly altered traditional views that life on this planet relies solely on photosynthetic energy (1, 2). Chemoautotrophic microbes that depend on sulfide-rich effluents constitute the base of the vent food chain (3). Free-living chemoautotrophs are grazed from surfaces or filtered from the water by a variety of animals, and symbiotic bacteria are found in the tissues of several vent-endemic invertebrates. Long term obligatory associations between symbiotic organisms and their hosts can result in the evolution of reciprocal adaptations or coadaptation (4). In addition to coadaptation, cospeciation also is considered strong evidence for coevolution (4). Cospeciation may be seen in the parallel cladogenic patterns of taxa involved in “tight” symbiotic or parasitic associations (5–7). To date, however, cospeciation patterns have not been found in the symbiotic bacteria associated with hydrothermal vent mollusks or vestimentiferan tubeworms (8–11). Herein, we report the first evidence for cospeciation between chemoautotrophic proteobacteria and their invertebrate hosts.
Symbiosis between chemoautotrophic bacteria and invertebrate animals first was described for vestimentiferan tubeworms from hydrothermal vents (12, 13). Other symbioses subsequently were reported for invertebrate animals from a variety of sulfide- or methane-rich marine environments (14). Clams of the family Vesicomyidae, which harbor intracellular bacteria in their gills (15), are found at hydrothermal vents and cold water sulfide/hydrocarbon seeps throughout the world’s oceans (16, 17). Evidence for coadaptation between these clams and their sulfur-oxidizing bacteria rests on the following observations: (i) The endosymbionts have not been found living freely in the environment (18, 19); and (ii) attempts to culture these endosymbionts have failed (14). Points i and ii constitute negative evidence and by themselves do not rule out a free-living stage for the bacteria. (iii) The clams have specialized morphological and biochemical adaptations that support the sulfur-oxidizing bacteria (15, 20–22); (iv) isotopic signatures from clam tissues clearly show chemoautotrophic carbon fixation (23); and (v) cytological evidence and molecular markers reveal that the bacterial symbionts are transmitted vertically within eggs (24, 25). Points iii–v are consistent with coadaptation and an obligatory relationship but do not rule out occasional acquisition of new symbionts, horizontal transmission of symbionts between clams, or host shifting between clam species. Similar degrees of coadaptation with chemoautotrophic symbionts are seen in lucinid clams, solemyid bivalves, and vestimentiferan tubeworms, but cospeciation has not been demonstrated within any of the host taxa (9–11), although specific bacterial associations exist at the level of host phyla (8).
Evidence for cospeciation lies in the pattern and timing of cladogenic (i.e., splitting) events in the associated taxa. A speciation event in the host lineage results in isolation and concomitant splitting of a vertically transmitted symbiont lineage. To test for cospeciation, the phylogenetic trees can be reconstructed separately for the host and symbiont lineages and then compared. If cospeciation is occurring, the topologies (i.e., branching order) of the trees should be parallel and the timing of splitting events should be approximately equivalent for the host and symbiont lineages. By contrast, host and symbiont trees may become independent if the symbionts occasionally are transferred horizontally between host individuals, recaptured from the natural environment, or switched between host species.
MATERIALS AND METHODS
Specimens.
We obtained specimens of nine vesicomyid clam species from a broad geographic range (Table 1). The species have been characterized in studies that used multi-locus allozymes and mitochondrial cytochrome oxidase subunit I (COI) sequences (17, 26). Taxonomic designations within the family Vesicomyidae are problematic due to the presence of morphologically “cryptic” species, phenotypic plasticity, and convergent morphological evolution (17, 26, 27). It should be noted that the present genus designations (Vesicomya, Calyptogena, and Ectenagena) do not reflect phylogenetic relationships. To avoid taxonomic ambiguity, we compared clam and symbiont DNAs isolated from the same individual of each host operational taxonomic unit). DNA sequences were obtained from at least two individuals of each host operational taxonomic unit.
Table 1.
Specimens, collection sites, and GenBank accession numbers
| Species | n | Latitude; longitude | Date | Dive* | Depth, m | Symbiont
|
Host
|
|
|---|---|---|---|---|---|---|---|---|
| 16S | COI | 16S | ||||||
| Calyptogena elongata | 3 | 34°N; 120°W | Dec. ’88 | dredge | 500 | AF035719 | AF008274 | AF035728 |
| C. kilmeri | 2 | 44°N; 125°W | 16 Jul. ’94 | A2796 | 795 | AF035720 | AF035941 | AF035729 |
| C. magnifica | 2 | 18°S; 113°W | Dec. ’93 | Nna12 | 2700 | AF035721 | AF008272 | AF035730 |
| C. n.sp. (Florida) | 2 | 26°N; 84°W | 3 Jun. ’92 | A2542 | 3313 | AF035722 | AF008281 | AF035731 |
| C. pacifica | 3 | 47°N; 129°W | 20 Jul. ’91 | A2413 | 2200 | AF035723 | AF008295 | AF035732 |
| C. phaseoliformis | 3 | 40°N; 144°E | 19 Jul. ’95 | S272 | 6370 | AF035724 | AF008283 | AF035733 |
| Ectenagena extenta | 2 | 41°N; 127°W | 30 Sep. ’90 | A2042 | 3271 | AF035725 | AF008266 | AF035734 |
| Vesicomya gigas | 3 | 27°N; 111°W | 26 Feb. ’93 | T2/26/93 | 2000 | AF035726 | AF008264 | AF035735 |
| V. lepta | 2 | 36°N; 122°W | 30 Dec. ’93 | V93-364-2 | 600 | AF035727 | AF035942 | AF035736 |
First letter indicates the submersible: A, Alvin; N, Nautile; S, Shinkai 6500; T, Turtle; V, Ventana.
PCR and Sequencing.
The clam hosts were examined for two mitochondrial genes [COI and large subunit (LSU) 16S rDNA]. The corresponding symbionts were examined for the small subunit (SSU) 16S rDNA. All host mitochondrial LSU and bacterial SSU sequences are new, and all but two of the host COI sequences have been reported previously (17). Nucleic acids were extracted by standard protocols from gill tissues of at least two individuals of each host species. Host mitochondrial COI was amplified and sequenced with primers and techniques that were described previously (17). Host mitochondrial LSU rDNA was amplified with the 16Sar and 16Sbr primers (28) under the same general conditions as COI, except for annealing at 50°C. Symbiont SSU rDNA was amplified with the 27F and 1492R primers under previously described conditions (9). Amplification products were precipitated with ammonium acetate:ethanol (0.5:2 volumes) and were used as templates for ABI Prism DNA sequencing reactions (Perkin–Elmer ABI). Sequencing primers VesLCO and VesHCO were used for COI; 16Sar and 16Sbr were used for mitochondrial LSU rDNA; and 27F, 344F, 516R, 530F, 884F, 1096R, and 1492R were used for bacterial SSU rDNA.
Phylogeny Reconstruction and Tests for Cospeciation.
Nucleotide sequences were aligned with clustalw (29), checked manually, and then used to examine evolutionary relationships within host clams and their symbiotic bacteria. The aligned data are available from the authors (A.S.P.) on request. By using the observed nucleotide frequencies and transition/transversion ratio, we applied the fastdnaml 1.0.6 program (30) or the dnaml program from phylip 3.57 (31), with randomized input of taxa (20 times) and global rearrangements. Bootstrap percentages from 1,000 resamplings were given above each node. Both trees were rooted with an appropriate outgroup. Each phylogeny was rooted with six different taxa, either heterodont clams or γ-subdivision proteobacteria, where outgroup and midpoint rooting all produced identical placement of the root node (not shown). Phylogenetic methods of distance, parsimony, and likelihood were used to reconstruct phylogenies without differences in the resulting topologies. Host and symbiont trees were compared under alternate data sets by Kishino–Hasegawa (KH) criteria (32). User-defined trees were submitted to fastdnaml and dnaml to estimate the SD in likelihood analysis. Phylogenetic topology comparisons considered topology only and did not constrain branch lengths between trees.
Phylogenetic Signal, Alternate Topologies, and Rate Estimates.
Phylogenetic signal within each data set was examined by calculating the skew (g1) in the logelikelihood (LnLi) distribution for all possible nine taxa topologies with paup*4v63 (unpublished paup* beta release v63; results published with permission of the author, D. L. Swofford). Phylogenies <40 LnLi’s from the maximum likelihood (ML) tree under each data set were saved from an exhaustive search, and topologies >40 LnLi’s were generated randomly with paup* and compared with the ML trees by KH criteria in paup*. The model of sequence evolution used was HKY85 + Γ with empirically estimated κ and α for each data set. All tests of phylogenetic hypotheses and the subsequent LnLi’s presented do not include outgroup taxa. Rates of molecular evolution were used to calculate the time of a last common ancestor by the following method: the maximal divergence within a group multiplied by 1/2 to generated the divergence per lineage then multiplied by a rate.
RESULTS
To test for cospeciation, we obtained host and symbiont DNA sequences from the muscle and gill tissues of individual clams. We examined portions of two mitochondrial genes from the clams [516 bp COI and 513 bp of LSU (16S) ribosomal DNA (LSU rDNA)] and 1,433 bp of the small subunit (16S) ribosomal DNA from the bacteria. Intraspecific sequence variation at the clam COI locus was <2%, and interspecific divergence exceeded 5%. Similarly, intraspecific variation at the clam LSU 16S locus was <1%, and interspecific divergence exceeded 4%. The bacterial SSU rDNA sequences revealed no intraspecific variation whereas interspecific divergence ranged from 0.9 to 3.9%.
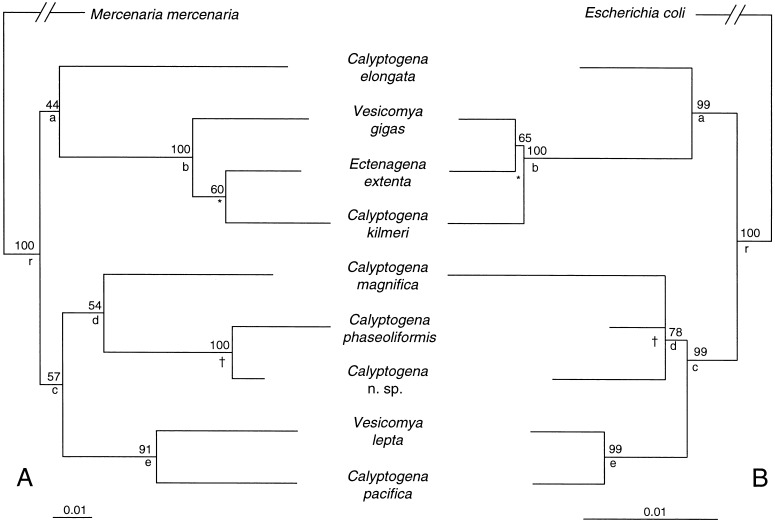
The DNA sequences were used separately to generate ML phylogenies for both the hosts and symbionts (Fig. 1). The clam COI and LSU rDNA sequences provided nearly identical phylogenetic topologies (not shown) and thus were combined for subsequent phylogenetic analyses. Five of the seven internal nodes (Fig. 1 a-e) were identical in the host and symbiont ML trees. One of the unshared internal nodes (∗) results from alternative resolutions of three closely related taxa. The second unshared node (†) results from a lack of resolution within the symbiont clade. The probability that such a match between host and symbiont phylogenies occurred by chance is low (P < 0.0001), assuming that the tree nodes were distributed independently and binomially (33). Adding appropriate outgroup taxa to each tree provides an additional node (r), rooting the host and symbiont trees in identical locations and decreasing the chance occurrence of a match between these two topologies (P < 5 × 10−6).
Figure 1.
ML phylogenies for nine species of vesicomyid clams and their associated endosymbionts. The clam species were characterized in previous allozyme and mitochondrial COI studies (17, 26). Host and symbiont trees were not drawn to the same rate scale (scale below each tree). Numbers above nodes are the percentage bootstrap support from 1,000 resamplings. Small letters and symbols below nodes mark topological comparisons between the two trees. All LnLi values presented are based on unrooted 9 taxa topologies. (A) The vesicomyid tree was based on combined data from portions of the mitochondrial COI gene (516 bp) and large subunit 16S rDNA (513 bp) gene, and an empirically derived ts/tv ratio of 3.0. (B) The bacterial tree (LnLi −2808.31) was based on a portion of small subunit 16S rDNA (1,433 bp), and an empirically derived ts/tv ratio of 3.75.
To test whether host and symbiont ML phylogenies were congruent, we estimated the likelihood of obtaining the symbionts’ tree topology given the host sequence data and vice versa (32). To do this, we constrained the ML topology of the bacteria on the clam sequences and estimated a new likelihood value (LnLi = −3346.68) that was compared with the ML tree (LnLi = −3344.15) for the clams alone (Table 2). The difference between the constrained and ML values (2.53) was not statistically significant. In other words, the symbiont topology was not significantly worse than the clam topology for explaining evolutionary relationships among the clam DNA sequences. The reciprocal test (clam topology constraining the bacterial data) also revealed no significant difference (LnLi = −2809.78; LnLi = −2808.31; difference = 1.47). Host and symbiont phylogenetic topologies were not significantly different.
Table 2.
Likelihood analyses based on phylogenetic hypotheses (H) and the observed DNA sequence data (R)
| Phylogenetic hypothesis, H | Sequence, R | Likelihood model | LnLi value | SD |
|---|---|---|---|---|
| ML clam | Clam | L(HC|RC) | −3344.15 | — |
| ML bacteria | Bacteria | L(HB|RB) | −2808.31 | — |
| Ho: clam and bacterial tree topologies were not different | ||||
| Bacterial tree topology | Clam | L(HMB|RC) | −3346.68 | 7.31 |
| Clam tree topology | Bacteria | L(HMC|RB) | −2809.78 | 2.17 |
| Ho: Phylogenies do not differ from expectations of a constant molecular clock | ||||
| Clock-like clam phylogeny | Clam | L(HMKC|RC) | −3349.69 | 3.16 |
| Clock-like bacterial phylogeny | Bacteria | L(HMKB|RB) | −2816.83 | 3.86* |
The model L(H|R) is read as the likelihood of hypothesis H given sequence data R, with the following subscripts: (C) clam; (B) bacteria; (M) maximum likelihood topology; and (MK) clock-like maximum likelihood phylogeny. The estimated standard deviation (S.D.) was obtained by the Kishino-Hasegawa method (32).
Significantly different from the appropriate ML comparison (α = 0.05).
A failure to reject a null hypothesis could be due either to data with insufficient signal or to limited statistical power. One limiting factor in phylogenetic hypothesis testing is the presence of significant signal within a given data set. The nucleic acid data from both host (g1 = −1.11) and symbiont (g1 = −1.33) contain significant phylogenetic signal (34). A second limiting factor is the power to reject alternate hypotheses under the KH criterion. We tested the power of the KH criterion to reject alternate topologies by retaining all topologies within 40 LnLis of each ML tree, resulting in 655 trees from the host and 675 from the symbiont. Topologies were removed from further consideration if they were rejected by KH criterion under either (i) their own data set or (ii) the alternate data set. This method retained the same nine topologies for each of the host and symbiont topology sets. These nine topologies differed at either one or two internal nodes. None of the 10,000 randomly sampled topologies >40 LnLis from the ML trees were retained by these criteria. Therefore, of the 1.35 × 105 possible topologies, we were able to reject reciprocally all but the same nine topologies with our method. All comparisons between these nine topologies are statistically significant and are consistent with the cospeciation hypothesis. These nine topologies were “insignificantly different” from each other by reciprocal KH tests but were distinguished by raw LnLi scores, and the best two are presented in Fig. 1.
The timing of cladogenic events in vertically transmitted symbionts should correspond roughly to those of the host. We were unable to compare the timing of internal cladogenic events because significant rate heterogeneity existed among the bacterial lineages (Table 2); however, divergence rates were not significantly heterogeneous among the clam lineages. Some fossil and molecular evidence suggest that vesicomyid clams diversified in the Cenozoic, during the last 50 million years (17), but additional fossil evidence suggests that vesicomyids extend to the Cretaceous, during the last 100 million years (35). If we assume bacterial 16S rDNA sequences diverge at a rate of 1–2% per 50 million years (5, 36), the maximal diversity observed among symbiont lineages (3.9%) results in estimated ages of 50–100 million years ago. Rate heterogeneity within the bacterial lineage suggests that ages based on the maximal observed diversity overestimate the age of the last common ancestor for the symbiont lineage. Thus, the estimated age of the bacterial symbiont lineage is probably <100 million years ago.
DISCUSSION
The present evidence for cospeciation between the vesicomyid clams and their vertically transmitted bacterial symbionts is strong. Cladogenic events of the hosts and symbionts are congruent. Furthermore, estimated ages of the host and symbiont clades were roughly comparable despite the uncertainties of molecular clock calibrations and the problem of internal rate heterogeneity among these bacteria. This result contrasts markedly with a parallel study of vestimentiferan tubeworm endosymbionts, which are not transmitted vertically (9, 10, 37). Evolutionary divergence among tubeworm endosymbionts preceded divergence of the vestimentiferan hosts by almost 200 million years (9).
The null hypothesis being tested herein is that host and symbiont phylogenetic topologies are congruent. A statistical problem with this hypothesis (and, for that matter, all hypotheses) is that failure to reject the null hypothesis does not prove the null is correct. However, we provide evidence that our data contain phylogenetic signal and further show that the method is able to differentiate among the vast majority of alternate hypotheses. Also, showing congruence between host and symbiont phylogenies does not necessitate that these arose by the mechanism we suggest, cospeciation. There are many ways that these phylogenetic patterns could be produced, but it certainly involves some long term and nonrandom association between host and symbiont. Considering the biological coadaptations in both host and symbiont and a lack of evidence for other processes that produce similar patterns, we propose cospeciation as a sufficient mechanism.
Given the many ways to obscure patterns of cospeciation between biologically associated lineages, the present results are remarkable. The lack of cospeciation between vestimentiferan tubeworms and the chemoautotrophic symbionts on which they depend completely for nutriment probably results from environmental acquisition of the bacteria in each generation (9, 10, 37). Similarly, cospeciation does not appear to occur in lucinid (38) or solemyid bivalves (11, 38). However, robust phylogenies are lacking for these bivalves, and lucinids may acquire endosymbionts environmentally (39). In contrast to the chemoautotrophic symbioses from the deep sea, cospeciation between heterotrophic bacteria and their insect hosts has been proposed in several associations, including the symbionts of aphids, cockroaches, tsetse flies, and carpenter ants (5, 40–42). However, robust phylogenies for most of the insect hosts are also not available (but see ref. 43).
If vesicomyid endosymbionts are transmitted strictly through the eggs, the evolutionary dynamics of host mitochondrial and endosymbiont genes should result in a close coupling. The present data from host mitochondrial sequences and symbiont sequences are therefore consistent with the hypothesis of nearly complete vertical transmission over a long term association. Lineage sorting or rare switching events between very closely related clam lineages may explain the minor discrepancies in the present topologies. Alternatively, the addition of more sequence information from hosts and symbionts may remove these discrepancies and more closely conform to strict vertical transmission. These data provide the first molecular evidence from both the hosts and symbionts for cospeciation between invertebrate animals and chemoautotrophic bacteria.
Acknowledgments
This is Contribution 98-21 of the Institute of Marine and Coastal Sciences, Rutgers University and New Jersey Agricultural Experiment Station Publication No. D-32104-3-98, supported by State funds and by National Science Foundation Grants OCE-89-17311, OCE 92-17026, OCE-93-02205, and OCE-96-33131 and National Institutes of Health Grant PHSTW00735-01 to R.C.V. and R.A.L.
ABBREVIATIONS
- COI
cytochrome oxidase subunit I
- LSU
large subunit
- SSU
small subunit
- KH
Kishino-Hasegawa
- LnLi
logelikelihood
- ML
maximum likelihood
Footnotes
Data deposition: See Table 1.
References
- 1.Lonsdale P. Deep Sea Res. 1977;24:857–863. [Google Scholar]
- 2.Ballard R D. Oceanus. 1977;20:35–44. [Google Scholar]
- 3.Jannasch H W, Wirsen C O. Bioscience. 1979;29:592–602. [Google Scholar]
- 4.Futuyma D J, Slatkin M. Coevolution. Sunderland, MA: Sinauer; 1983. p. 555. [Google Scholar]
- 5.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc Lond B. 1993;253:167–171. [Google Scholar]
- 6.Cunningham C W, Buss L W, Anderson C. Evolution. 1991;45:1301–1315. doi: 10.1111/j.1558-5646.1991.tb02637.x. [DOI] [PubMed] [Google Scholar]
- 7.Hafner M S, Nadler S A. Nature (London) 1988;332:258–259. doi: 10.1038/332258a0. [DOI] [PubMed] [Google Scholar]
- 8.Distel D L, Felbeck H, Cavanaugh C M. J Mol Evol. 1994;38:533–542. [Google Scholar]
- 9.Feldman R A, Black M B, Cary C S, Lutz R A, Vrijenhoek R C. Mol Mar Biol Biotech. 1997;6:268–277. [PubMed] [Google Scholar]
- 10.Laue B E, Nelson D C. Mol Mar Biol Biotech. 1997;6:180–188. [PubMed] [Google Scholar]
- 11.Krueger D M, Cavanaugh C M. Appl Environ Microbiol. 1997;63:91–98. doi: 10.1128/aem.63.1.91-98.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavanaugh C M, Gardiner S L, Jones M L, Jannasch H W, Waterbury J B. Science. 1981;213:340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 13.Felbeck H. Science. 1981;213:336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 14.Nelson D C, Fisher C R. The Microbiology of Deep-Sea Hydrothermal Vents. Boca Raton, FL: CRC; 1995. pp. 125–167. [Google Scholar]
- 15.Fiala-Medioni A, LePennec M. Oceanol Acta. 1988;11:185–192. [Google Scholar]
- 16.Boss K J. Malacologia. 1969;9:254–255. [Google Scholar]
- 17.Peek A S, Gustafson R G, Lutz R A, Vrijenhoek R C. Mar Biol. 1997;130:151–161. [Google Scholar]
- 18.Polz M F, Cavanaugh C M. Proc Natl Acad Sci USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyer C L, Dobbs F C, Karl D M. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boss K J, Turner R D. Malacologia. 1980;20:161–194. [Google Scholar]
- 21.Childress J J, Fisher C R, Favuzzi J A, Arp A J, Oros D R. J Exp Biol. 1993;179:131–158. [Google Scholar]
- 22.Childress J J, Fisher C R, Favuzzi J A, Sanders N K. Physiol Zool. 1991;64:1444–1470. [Google Scholar]
- 23.Rau G H. Science. 1981;213:338–340. doi: 10.1126/science.213.4505.338. [DOI] [PubMed] [Google Scholar]
- 24.Endow K, Ohta S. Mar Ecol Prog Ser. 1990;64:309–311. [Google Scholar]
- 25.Cary S C, Giovannoni S J. Proc Natl Acad Sci USA. 1993;90:5695–5699. doi: 10.1073/pnas.90.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vrijenhoek R C, Schutz S J, Gustafson R G, Lutz R A. Deep Sea Res. 1994;41:1171–1189. [Google Scholar]
- 27.Kojima S, Seawa R, Kobayashi T, Hashimoto T, Fujikura K, Hashimoto J, Ohta S. Mar Biol. 1995;122:401–407. [Google Scholar]
- 28.Palumbi S R. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 205–247. [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen G J, Matsuda H, Hagstrom R. CABIOS. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 32.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 33.Penny D, Foulds L R, Hendy M D. Nature. 1982;297:197–200. doi: 10.1038/297197a0. [DOI] [PubMed] [Google Scholar]
- 34.Hillis D M, Huelsenbeck J P. J Heredity. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- 35.Kanie Y, Sakai T. VENUS. 1997;56:205–220. [Google Scholar]
- 36.Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 37.Cary S C, Warren W, Anderson E, Giovannoni S J. Mol Mar Biol Biotech. 1993;2:51–81. [PubMed] [Google Scholar]
- 38.Durand P, Gros O, Frenkiel L, Prieur D. Mol Mar Biol Biotech. 1996;5:37–42. [Google Scholar]
- 39.Gros O, Darrasse A, Durand P, Frenkiel L, Moueza M. Appl Environ Microbiol. 1996;62:2324–2330. doi: 10.1128/aem.62.7.2324-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandi C, Sironi M, Damiani G, Magrassi L, Nalepa C A, Laudani U, Sacchi L. Proc R Soc Lond B. 1995;259:293–299. doi: 10.1098/rspb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 41.Askoy S, Pourhosseini A A, Chow A. Insect Mol Bio. 1995;4:15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 42.Schröder D, Deppisch H, Obermayer M, Krohne G, Stackebrandt E, Hölldobler B, Goebel W, Gross R. Mol Micro. 1996;21:479–489. doi: 10.1111/j.1365-2958.1996.tb02557.x. [DOI] [PubMed] [Google Scholar]
- 43.Moran N A, von Dohlen C D, Baumann P. J Mol Evol. 1995;41:727–731. doi: 10.1007/BF00170675. [DOI] [PubMed] [Google Scholar]