Figure 1.
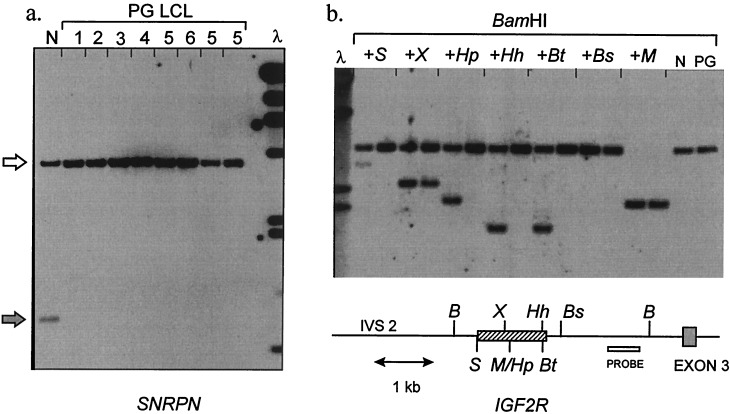
Uniparental methylation pattern at imprinted loci in human parthenogenetic LCL. (a) Methylation of SNRPN (chromosome 15q11-q13). The probe used for Southern blotting is KB17, a standard diagnostic reagent for Prader–Willi syndrome. DNAs are digested with XbaI + NotI. The paternal allele (gray arrow) is unmethylated, yielding a 0.9-kb fragment from normal DNA (N). 1–6, six subcultured lines of parthenogenetic lymphoblastoid cells (LCL) established from patient F.D. Only the methylated 4.2-kb maternal band (white arrow) is present. λ, λHindIII marker; 23, 9.4, 6.6, 4.4, 2.3, 2.0, 0.56 kb. (b) Methylation of IGF2R. The probe (open box) is fragment Bx of clone pE3UP (25), which detects a 3.2-kb BamHI fragment. Pairs of lanes contain normal (N) or parthenogenetic (PG) LCL DNA digested with BamHI, alone or plus a second enzyme as indicated: B, BamHI; S, SacII; X, XhoI; M/Hp, MspI/HpaII; Hh, HhaI; Bt, BstUI; Bs, BssHII. For each enzyme only the site nearest the probe is shown. The cross-hatched box indicates the region of allele-specific methylation; the HpaII, HhaI, and BstUI sites are methylated only on a maternal allele; the SacII site in addition is methylated incompletely on a paternal allele; the XhoI is methylated incompletely on a maternal allele and unmethylated on a paternal allele. These results concur with previous data (25) and indicate faithful allele-specific methylation across this region in the PG LCL DNA. λ, λHindIII marker; 6.6, 4.4, 2.3, 2.0 kb.