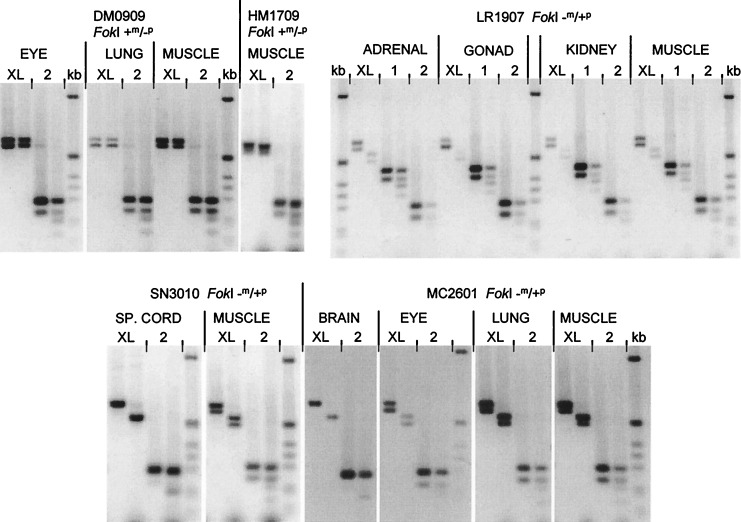
Figure 4.
Exclusively paternal expression of XLαs-containing GNAS1 transcripts. All experiments used a downstream PCR primer in exon 6; the location of the upstream primer is indicated (either in the XLαs exon, exon 1 or exon 2) above each pair of lanes. Each pair of lanes contains undigested (Left) or FokI-digested (Right) RT-PCR product. The two bands seen in most undigested samples result from alternative splicing of exon 3. In fetuses DM0909 and HM1709, the paternal allele is lacking a FokI site (genotype +m/−p). The XLαs-derived RT-PCR products (XL) are completely FokI-resistant. The total GNAS1 transcripts, in contrast (2), are partially cleaved by FokI, indicating their biallelic origin. In fetus LR1907, the genotype is −m/+p. The XLαs-derived products are digested to completion by FokI, again demonstrating their exclusively paternal origin. Also, in this panel, exon 1-specific transcription has been analyzed separately, showing biallelic expression from the promoter upstream of exon 1. Fetuses SN3010 and MC2601 are again FokI −m/+p. RT-PCR products are digested to completion if XLαs-derived but only partly when the exon 2 primer is used. kb, 1-kb ladder: 1,018, 517/506, 396, 344, 298, 220, 201, 154, 134 bp.