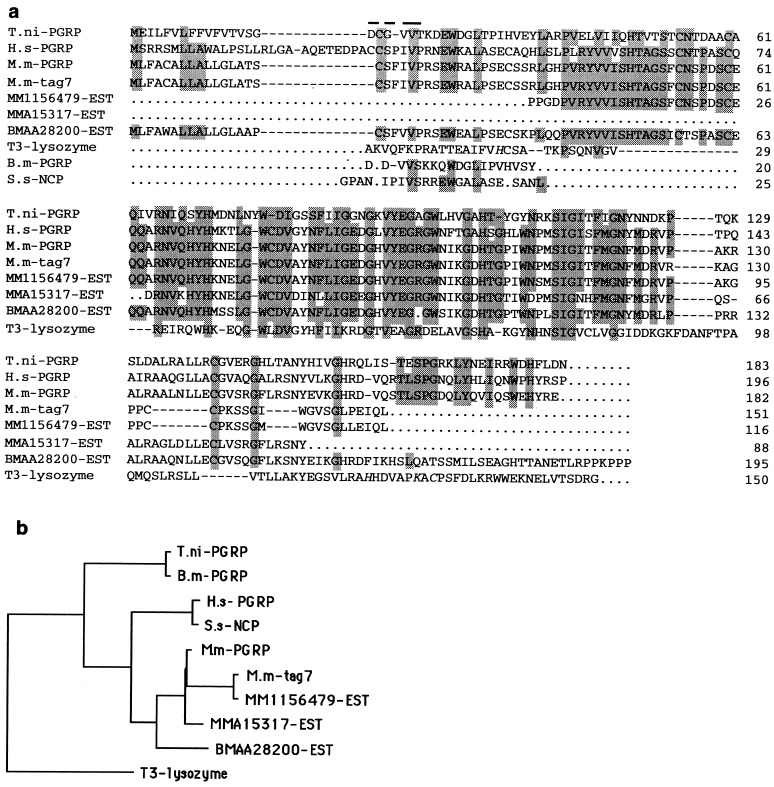
Figure 1.
(a) Deduced amino acid sequence of T. ni PGRP, murine M.m-PGRP, and human H.s-PGRP cDNA (GenBank accession nos. AF076481, AF076482, and AF076483). Homologs found by blast and fasta database searches (29) were M.m-tag7 (X86374) from mouse; MM1156479 and MMA15317, both expressed sequence tags (ESTs) from mouse; BMAA28200 from the nematode Brugia malayi; T3 lysozyme (P20331) from phage T3; B.m-PGRP (1839611) from Bombyx mori; and S.s-NCP (P80552) from pig neutrophils. Determined or predicted (15) signal peptides are separated from the mature proteins by a gap (−). Missing sequence data are indicated by periods. Conserved residues in all but one of the given sequences (excluding that of T3 lysozyme) are printed on gray background. Residues obtained by amino acid sequencing of the baculovirus-expressed T. ni PGRP are overlined. The T3 lysozyme active site residues (His-17, Tyr-46, His-122, Lys-128, and Cys-130) are shown in italics. (b) Phylogenetic tree derived from the amino acid sequences by using the branch-and-bound algorithm of the paup program (20). Branch lengths are shown proportional to the minimal number of changes.