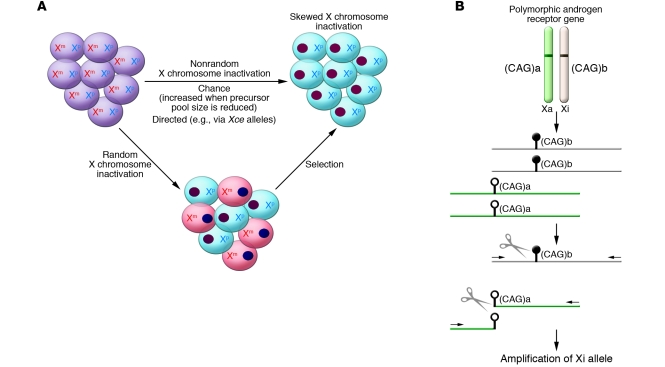
Figure 1. Skewed XCI in females.
(A) Skewed XCI can result from either a primary nonrandom inactivation or a secondary, acquired nonrandom inactivation. There are two active X chromosomes in the female embryo, one from each parent (maternal and paternal; Xm and Xp, respectively). Upon inactivation, one X forms the heterochromatic inactive X depicted here as a darkly staining oval. The cells are shaded to reflect the different origins of their remaining active X. Subsequent stochastic or selective growth of one cell population leads to skewing of inactivation. However, if the initial inactivation was influenced by an allele (analogous to the murine Xce allele) then inactivation could be directed primarily towards one allele. A reduction in the embryonic precursor pool size at the time of random choice would make skewed XCI more likely. (B) The common way to examine XCI skewing makes use of a highly polymorphic CAG repeat (shown here as having allele sizes a and b) in the androgen receptor gene and differential methylation (methylation shown as black lollipop while the unmethylated allele has open lollipops) of the active (Xa) and inactive (Xi) X. DNA is extracted from the cells to be tested and then digested with a methylation-sensitive restriction enzyme (shown as scissors). The methylated (Xi) DNA will remain intact for subsequent PCR amplification using primers (shown as arrows) flanking the cut site. The ratio of the alleles (a and b, distinguished by PCR product length) in the undigested compared with the digested DNA reflects the XCI skewing.