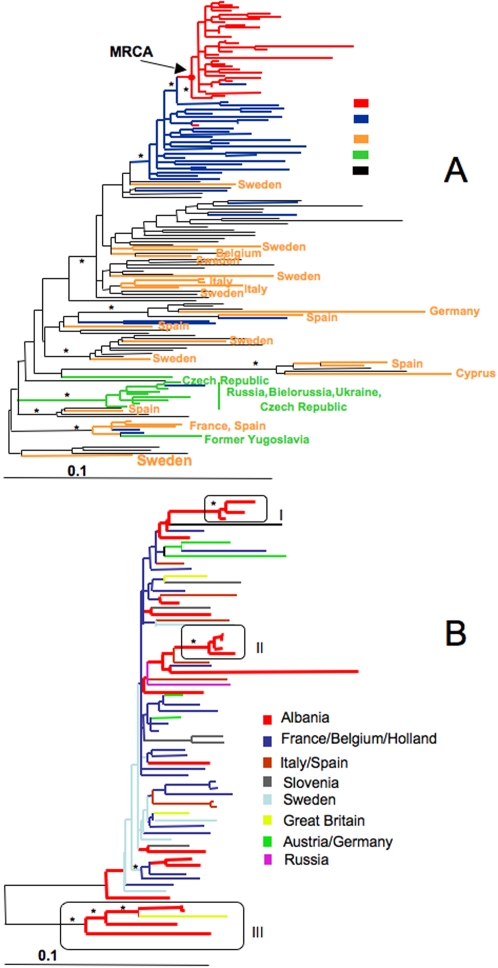
Figure 1. Maximum likelihood phylogenetic analysis of HIV-1A pol sequences.
Branch lengths were estimated with the best fitting nucleotide substitution model according to a hierarchical likelihood ratio test [29], and were drawn in scale with the bar at the bottom indicating 0.1 nucleotide substitutions per site. One * along a branch represents significant statistical support for the clade subtending that branch (p<0.001 in the zero-branch-length test and bootstrap support >75%). The color of each tip branch represents the country (or geographic region) of origin of sequence corresponding to that branch, according to the legend in the figure. A. Phylogenetic tree of 31 HIV-1A strains from Bulgaria and 122 subtype A reference sequences downloaded from the Los Alamos HIV database. The tree was rooted using two HIV-1B strains as outgroup. The arrow indicates the most recent common ancestor (MRCA) of the Albanian monophyletic clade. B. Phylogenetic tree of 21 HIV-1B strains from Bulgaria and 46 subtype B reference sequences downloaded from the Los Alamos HIV database. The tree was rooted using two HIV-1A strains as outgroup. Solid boxes highlight statistically supported clades of Albanian sequences.