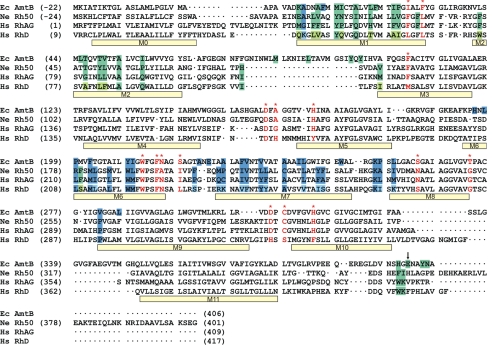
Fig. 3.
Structure-based sequence alignment of NeRh50 and EcAmtB. The program Sequoia (58) was used to carry out the structure-based alignment and combine it with a sequence-based alignment for HsRhAG and RhD. The alignment before the start of M1 was adjusted manually. Other minor adjustements were made in the structurally nonhomologous parts of loops. The vertical arrow at the CTR marks the position beyond which the alignment is not meaningful, and the sequences are merely shown for completeness. Negative sequence numbers are assigned to the processed leader peptides of EcAmtB and NeRh50, hence the mature proteins as expressed in E. coli start at residue +1. Transmembrane helices M1-M11 as observed in the NeRh50 structure and M0 as predicted (23) are indicated with bars below the aligned sequences. Residues with large interface contacts (see SI Fig. 8) are in boxes colored according to whether they belong to the green or dark blue subunit in Fig. 1. The corresponding residues in RhD are shown in lighter colors if different from those of RhAG. Selected residues along the substrate pathway as discussed in the text are marked in red.