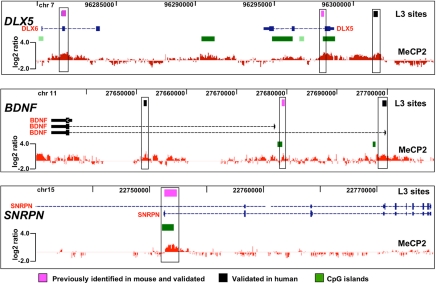
Fig. 2.
Validation of ChIP–chip analysis on established MeCP2-binding sites. A representative histogram of MeCP2 log2 signal ratio values is shown below the University of California, Santa Cruz, Genome Browser window containing known genes, CpG islands (green boxes), and L3 sites (black and pink boxes) for each MeCP2 target gene locus. For DLX5 and DLX6 controls, one site was previously identified in mouse (right pink box) (19) and validated (SI Fig. 7), and another site in DLX6 (left pink box) was also identified in mouse and validated by conventional ChIP analysis (19). For BDNF, an MeCP2 site in the first intron (pink box) was observed at the same position as previously shown in mouse and rat Bdnf (21, 22) and appears to control activation-induced transcription of the gene in neurons (44). Additional new MeCP2 sites were observed and validated (black boxes). A site in SNRPN that serves as the paternal imprinting control region for 15q11–13 was included (pink box) as an additional control. Analyses of this site in both mouse and human by conventional ChIP revealed binding by MeCP2 (16, 17). However, binding of MeCP2 to this site does not affect transcription of SNRPN (16, 19). The SNRPN site was validated by ChIP–chip at levels L1–L4.