Abstract
Mycoplasma mobile, a parasitic bacterium lacking a peptidoglycan layer, glides on solid surfaces in the direction of a membrane protrusion at a cell pole by a unique mechanism. Recently, we proposed a working model in which cells are propelled by leg proteins clustering at the protrusion's base. The legs repeatedly catch and release sialic acids on the solid surface, a motion that is driven by the force generated by ATP hydrolysis. Here, to clarify the subcellular structure supporting the gliding force and the cell shape, we stripped the membrane by Triton X-100 and identified a unique structure, designated the “jellyfish” structure. In this structure, an oval solid “bell” ≈235 wide and 155 nm long is filled with a 12-nm hexagonal lattice and connected to this structure are dozens of flexible “tentacles” that are covered with particles of 20-nm diameter at intervals of ≈30 nm. The particles appear to have 180° rotational symmetry and a dimple at the center. The relation of this structure to the gliding mechanism was suggested by its cellular localization and by analyses of mutants lacking proteins essential for gliding. We identified 10 proteins as the components by mass spectrometry and found that these do not show sequence similarities with other proteins of bacterial cytoskeletons or the gliding proteins previously identified. Immunofluorescence and immunoelectron microscopy revealed that two components are localized at the bell and another that has the structure similar to the F1-ATPase β subunit is localized at the tentacles.
Keywords: bacteria, electron microscopy, gliding motility, immunofluorescence, protein identification
Mycoplasmas are commensal and occasionally parasitic bacteria with small genomes that lack a peptidoglycan layer (1). Several mycoplasma species form membrane protrusions, such as the head-like structure in Mycoplasma mobile and the attachment organelle in Mycoplasma pneumoniae (2–8, 46). On solid surfaces, these species exhibit gliding motility in the direction of the protrusion; this motility is believed to be involved in the pathogenicity of mycoplasmas (3–5, 9, 10, 46). Interestingly, mycoplasmas have no surface flagella or pili, and their genomes contain no genes related to known bacterial motility. In addition, no homologs of motor proteins that are common in eukaryotic motility have been found (3–5, 11, 46).
M. mobile, isolated from the gills of a freshwater fish in the early 1980s, is a fast-gliding mycoplasma (12–16). It glides smoothly and continuously on glass at an average speed of 2.0 to 4.5 μm/s, or three to seven times the length of the cell per second, exerting a force of up to 27 piconewtons (pN). Recently, we identified huge proteins involved in this gliding mechanism (17–21), visualized the putative machinery and the binding protein (22, 23), and identified the direct energy source used and the direct binding target (24–26). On the basis of these results, we proposed a working model in which cells are propelled by “legs” composed of Gli349 repeatedly catching and releasing sialic acids fixed on the glass surface (3, 4, 18, 19, 27, 46) and driven by the force exerted by P42 (21) through Gli521 molecules, based on the energy of ATP hydrolysis.
This scenario leads to a crucial question: What physical structure could support a gliding force as strong as 27 pN at maximum, while maintaining the flask cell shape? As in the case of other mycoplasmas, M. mobile does not have a bacterial cell wall—i.e., a peptidoglycan layer. Moreover, the genome does not have bacterial cytoskeletal proteins, such as MreB or FtsZ (28, 29). M. pneumonia, which is positioned at some distance from M. mobile on the phylogenetic tree in mycoplasmas, also can glide by its membrane protrusion (3, 4, 10, 30, 46). This species has a cytoskeletal structure in the membrane protrusion, and some of its protein components have been identified (2–5, 31, 32, 46). The M. mobile genome also lacks any homologs of these protein components (11). In consideration of these facts, we currently have no clear answer to the above question.
In this study, to address this mystery, we analyzed the interior structures of M. mobile cells treated by a detergent under electron microscopy (EM), and found a cytoskeletal structure, plausibly supporting the gliding force of M. mobile.
Results
Surface and Inside Structures of the Cell Neck.
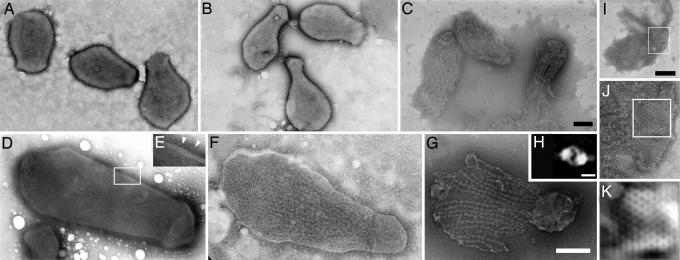
To determine the architecture of the gliding machinery, we observed the cell structures after treatment with various concentrations of Triton X-100 (Fig. 1). The EM images of negatively stained intact cells showed a conical cell shape (Fig. 1 A and D), consistent with our previous work (15, 18, 19, 33). On the cell surface, some filamentous structures were observed (Fig. 1E). However, we cannot trace the outlines of these structures or claim that they are restricted to a special part because of the insufficient contrast of the images.
Fig. 1.
Negative staining EM images of cells treated with different concentrations of Triton X-100. (A and D) Intact cells. (E) The field image indicated by the rectangle is magnified and adjusted for contrast to show the filamentous structures marked by white triangles. (B and F) Cells treated with 0.01% Triton X-100. (C and G) Cells treated with 0.1% Triton X-100. (A–C) Field images. (D, F, and G) Single-cell images. (Scale bars, 200 nm.) (H) Averaged image of the particles in jellyfish “tentacles.” Nine hundred seventy particle images from six jellyfish structures were integrated into one image. (Scale bar, 10 nm.) (I–K) An image of the jellyfish structure where the lattice of bell can be observed easily. The fields indicated by the rectangles in I and J are magnified in J and K, respectively. (K) The image was Fourier transformed, reduced of noise, and then reconstituted. (Scale bar, 200 nm.)
Next, we observed the cells treated with 0.01% Triton X-100 (Fig. 1 B and F). Under this condition, the cell membranes were partially damaged, and intracellular structures were observed, i.e., a solid block was apparent at the cell head, and many filaments could be seen at the neck. This structure, which we previously designated the “ghost,” can be reactivated for gliding when ATP is added extracellularly (26). We tried treatment with various concentrations of Triton X-100 and found that the appearance of the cells did not change under the range of 0.003% to 0.03% Triton X-100. When we treated the cells with 0.1% Triton X-100, the inside structure was clearly visible (Fig. 1 C and G). The architecture found here featured a solid oval or half sphere at the cell head, and this structure is connected with dozens of filaments, which are decorated with periodicity of 30 nm by particles ≈20 nm in diameter (Fig. 2). Based on this appearance, we named this cytoskeletal structure the “jellyfish” structure. The longer and shorter axes of the oval “bell,” which were taken as the mean value for 20 cells, were 235 ± 31 nm and 155 ± 32 nm. Occasionally, the bells show a lattice (Fig. 1 I and J). Fourier transformation of the bell images showed clear periodicity of ≈12 nm and hexagonal alignment (Fig. 1K). Although this lattice is likely derived from the intrinsic structure of bell, it is not always visible. The lattice may be an internal structure of the bell, and its appearance may depend on the staining condition of the jellyfish structure. The tentacles of the jellyfish structure were covered with particles. The number of 20-nm particles varied from 225 to 113 per cell. To clarify the outline of these particles by reducing the noise, we collected 970 particle images from six jellyfish structure images, and performed averaging. The images were integrated into the single image shown in Fig. 1H, which suggested a rotational symmetry of 180°. The particle appears to be formed of two identical blocks, each with two arms.
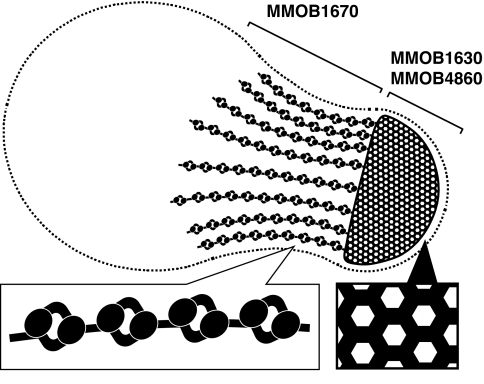
Fig. 2.
Schematic of the cytoskeletal structure of an M. mobile cell. A solid structure, the bell, is located at the cell head and filled with a hexagonal lattice of 12-nm periodicity. Dozens of tentacles are connected to the bell, and these tentacles are covered with particles of 20-nm diameter at intervals of ≈30 nm. The particles appear to have rotational symmetry through 180°.
In previous studies, we tried to identify the putative inside structure by Triton extraction but failed (19). In the present study, we improved two points. First, we treated the cells simultaneously by DNase. The removal of DNA strings greatly improved EM imaging and protein fractionation. Second, here, we used the adhesive strain, the gli521 mutant (P476R), as the standard strain (26). The adhesive character was advantageous to keep the jellyfish structures on the grid through the treatments, resulting in ≈5-fold numbers of jellyfish structures than the case of the wild-type strain. No apparent structural differences could be found on the jellyfish structures between the wild type [supporting information (SI) Fig. 7] and the adhesive strains.
Jellyfish Structure in Mutants Deficient in Gliding.
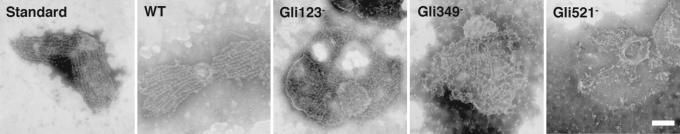
The localization of tentacles of the jellyfish structure corresponds to that of the gliding machinery, suggesting the involvement of gliding proteins in the jellyfish structure. Therefore, we compared the Triton-extracted structures of normally gliding strains and the nongliding and nonbinding mutants lacking each of the gliding proteins (Fig. 3). The bell of the jellyfish structure in the mutants appeared to be similar to that of the gliding positive strains. However, the continuity and numbers of tentacles were reduced in the mutants. Most significant changes were found in the m9 mutant, which lacked the Gli521 protein. This disorder may suggest that the jellyfish structure is supported by the machinery formed by the gliding proteins clustering at the cell neck, although the jellyfish structure is not composed of these proteins (see below).
Fig. 3.
Jellyfish structures of mutant cells lacking proteins that are involved in the gliding mechanism. The cells were treated by 0.3% Triton X-100 and adsorbed to grids. Strains are indicated at the upper left of each image. Standard, WT, Gli123−, Gli349−, and Gli521− represent the standard, the wild type, m12, m13, and m9 strains, respectively. (Scale bar, 200 nm.)
Identification of Protein Components of the Jellyfish Structure.
To identify the protein components of the jellyfish structure, we collected a Triton X-100-insoluble fraction of the cells and performed peptide mass fingerprinting (PMF). The cells were treated with various concentrations of Triton X-100, collected for the insoluble fractions, and subjected to SDS/PAGE (Fig. 4). The number of protein bands decreased with the increase in the Triton X-100 concentration used. When the cells were treated with 0.3% Triton X-100, the protein profile of the fraction showed nine major protein bands, and the jellyfish structures could be clearly observed under EM. Gli521, Gli349, and Gli123 proteins were removed from this fraction to be <2% of the original amount. Thus, we concluded that these gliding proteins are not involved in the jellyfish structure. We cut the nine protein bands from the SDS/PAGE, subjected them to PMF, and succeeded in identifying the ORFs for six of them (Table 1). We were unable to identify the ORFs for three of the proteins because of weak mass spectrometry signals or confusing results, which may have been caused by the mixture of source proteins. We further examined the five minor bands found in the fraction treated with 0.3% Triton X-100, because these proteins were suggested to interact with the jellyfish structure. To overcome the weakness of mass spectrometry signals caused by the low levels of protein, we used the corresponding protein bands from the trial using 0.03% Triton X-100 as a substitute. The protein bands a, e, h, and i could be clearly identified, indicating a lack of contamination by other proteins, showing that these protein bands were identical with the corresponding bands focused in the fraction that was treated with 0.3% Triton X-100. The fraction of the jellyfish structure from the wild-type strain showed the protein profiles indistinguishable from those of the standard strain, and the same ORFs were identified by mass spectrometry (data not shown).
Fig. 4.
Protein profiles of Triton X-100 insoluble fractions treated with various concentrations of Triton X-100. Fractions were subjected to SDS/PAGE and stained by Coomassie brilliant blue (CBB). (Left and Right) Gel images of 5.5% and 12.5% polyacrylamide, respectively. The amount of fraction applied to each lane was adjusted to derive from the same amount of mycoplasma culture. Twice the amount was applied to the 5.5% polyacrylamide gel than that to the 12.5% gel. The molecular mass and the band positions of gliding proteins previously identified are indicated on the left (20). Lane 1, the whole-cell lysate. Insoluble fractions after treatment by 0.01%, 0.03%, 0.1%, and 0.3% Triton X-100 are shown by lanes 2, 3, 4, and 5, respectively. Solid triangles marked a–j indicate protein bands that were identified by PMF. The a′ band is derived from the same protein as that of a. Open triangles indicate protein bands that could not be identified.
Table 1.
Proteins identified as components of the jellyfish structure
| Protein band* | Gene ID | Annotation | MW,† kDa |
|---|---|---|---|
| a | MMOB1650 | Unknown | 127.2 |
| b | MMOB1670 | F1-ATPase β-subunit | 88.4 |
| c | MMOB4530 | Phosphoglycerate kinase | 56.6 |
| d | MMOB1660 | F1-ATPase α-subunit | 58.7 |
| e | MMOB0150 | Xylose solute-binding protein | 56.9 |
| f | MMOB1630 | Unknown | 39.7 |
| g | MMOB1620 | Unknown | 33.7 |
| h | MMOB1640 | Unknown | 22.8 |
| i | MMOB5430 | Unknown | 18.2 |
| j | MMOB4860 | Unknown | 17.5 |
*The protein bands are indicated in Fig. 4.
†Molecular mass predicted from amino acid sequences.
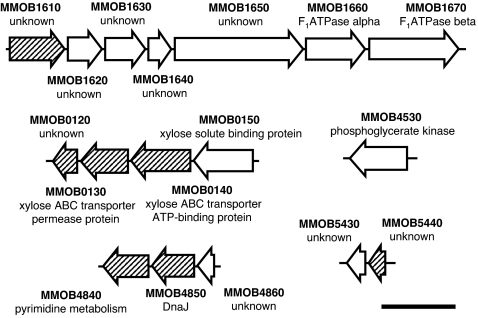
The component proteins are coded in five loci on the genome (Fig. 5). Six of the proteins are tandemly coded in a locus, as MMOB1620, MMOB1630, MMOB1640, MMOB1650, MMOB1660, and MMOB1670 from the 5′ end (11). Four of 10 identified ORFs have annotations, i.e., MMOB1660, MMOB1670, MMOB0150, and MMOB4530 are assigned to the F1-ATPase α-subunit, F1-ATPase β-subunit, xylose solute-binding protein, and phosphoglycerate kinase, respectively. MMOB1660 and MMOB1670 have clear similarity to a component of proton pump, namely F0F1 ATPase, but no similarity was found between neighboring ORFs and the other components of functional proton pumps (34).
Fig. 5.
ORFs in gene loci encoding protein components of the jellyfish structure. Open arrows indicate ORFs coding the components. (Scale bar, 2 kb.)
Subcellular Localization of Protein Components of the Jellyfish Structure.
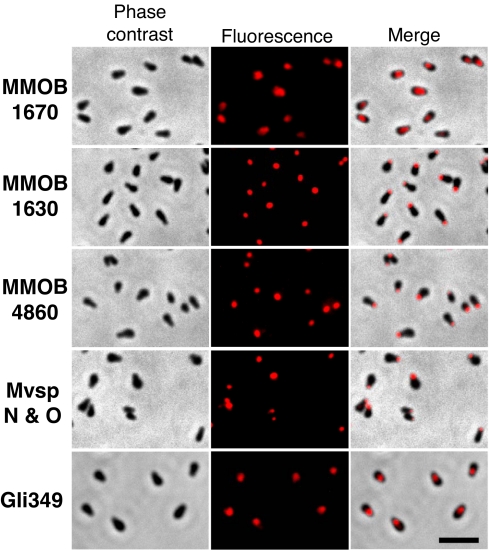
To determine the localization of these protein components of the jellyfish structure, we made polyclonal antibodies and performed immunofluorescence microscopy (Fig. 6). The antibodies against the bands of the protein encoded by MMOB1620, MMOB1630, MMOB1660, MMOB1670, and MMOB4860 were prepared in mice. Western blot analysis of the whole-cell lysate of M. mobile showed that these antibodies reacted only to the targeted proteins. Next, we performed immunofluorescence microscopy using these antibodies. No signal was detected when we fixed and labeled intact mycoplasma cells, as distinct from the results obtained for gliding proteins, including Gli349. These results are consistent with the EM observations, suggesting that the jellyfish structure lies within the interior of the cells. Therefore, we permeabilized the fixed cells with 0.1% Triton X-100 and labeled the proteins. The fluorescent signals clearly showed the localization of the proteins encoded by MMOB1630, MMOB1670, and MMOB4860 (Fig. 6). Previously, we reported that the cell surface of M. mobile can be divided into three parts, namely, the head, body, and neck (17). This feature can be easily observed by localization of surface proteins. All of the proteins involved in the gliding mechanism are localized at the neck, as represented by Gli349 (19), and the proteins possibly involved in antigenic variation are localized in other regions, as represented by mobile variable surface proteins (Mvsps) N and O (17) (Fig. 6). The localization of three proteins of the jellyfish structure corresponded to this differentiation. The MMOB1630 and MMOB4860 proteins were localized in the head, as observed for Mvsps N and O, and the MMOB1670 protein was in the neck, as observed for Gli349 (Figs. 2 and 6). Antibodies to the MMOB1620 and MMOB1660 proteins did not show any signal, even when higher concentrations were applied for labeling, although the antibodies reacted to the target proteins with a level of sensitivity similar to that shown by other antibodies. These results suggest that the parts recognized by the antibodies of these two proteins are occluded under the conditions used.
Fig. 6.
Subcellular localization of protein components of the jellyfish structure examined by immunofluorescence microscopy. The labeled proteins are indicated at the left by their gene IDs. Phase contrast, fluorescence and merged images are presented in Left, Center, and Right, respectively. The cells were chemically fixed, permeabilized by 0.1% Triton X-100, and labeled for the proteins other than Gli349. The permeabilizing step was not applied for labeling of Gli349. MMOB1670, MMOB1630, and MMOB4860 proteins correspond to the bands b, f, and j, respectively, shown in Fig. 4. The amino acid sequence of MvspN shared 314 aa with the MvspO sequence of 458 aa, and the antibody used here is known to recognize both proteins equally in Western blotting (17). (Scale bar, 2 μm.)
We also examined subcellular localization of MMOB1630, MMOB1670, and MMOB4860 by ImmunoGold EM and obtained the results consistent with those from immunofluorescence microscopy (SI Fig. 8). MMOB1630 and MMOB4860 did not show specific localization in the bell. The target of antiMMOB1670 could not be identified in the image of tentacles, because the chemical fixation needed for ImmunoGold staining reduced the image resolution.
Discussion
Structures on the Cell Neck Surface.
In a previous study, a leg structure of M. mobile was identified by quick freeze-fracture rotary-shadow EM (23). This structure, which was 50 nm long and 4 nm thick, protruded from the cell membrane and attached to the glass surface at its distal end. In the current study, the negative-staining EM method showed thin filamentous structures that were not found in previous studies (Fig. 1E) (10, 35). These structures and the legs found under freeze-fracture EM differed in terms of their stiffness and thickness. However, we suggest that these images may have been derived from the same structure for the following reasons. First, as the major portion of each cell was removed in the freeze fracture procedure, only the legs tightly bound to the glass surface with some tension could be observed. This may have also caused changes in the apparent stiffness. Second, generally, deep etching and rotary shadowing, which are required for freeze-fracture EM, tend to make the structures look thicker.
Possible Roles of the Jellyfish Structure in Gliding.
The cytoskeletal structure “jellyfish” (Fig. 2) is expected to play some role in the gliding mechanism for two reasons. First, the localization of tentacles corresponds to that of the gliding proteins (17–20), and second, the structure of the “jellyfish” was disordered in the mutants lacking gliding proteins, suggesting a physical interaction between the jellyfish structure and gliding machinery. In turn, what are the roles of the jellyfish structure in the gliding mechanism? Currently, we can propose three possibilities. Namely, the structure acts as (i) a scaffold to support the gliding machinery mechanically, (ii) a protein transporter to assemble the gliding machinery on the cell surface, or (iii) a force transmitter to coordinate leg movements. We examined the effects of antibodies against the component proteins of the jellyfish structure on gliding by using the gliding ghost system, in which the permeabilized cells can be reactivated for gliding by the addition of ATP (26). None of the antibodies had any observable effect, although the accessibility of these antibodies was confirmed by immunofluorescence microscopy of chemically fixed ghosts (data not shown). However, this result does not necessarily rule out the above possibilities, because the binding of antibodies cannot always inhibit the protein function.
Jellyfish Structure Is Internal.
The jellyfish structure was found by removing the cell membrane by Triton X-100 extraction, and none of the antibodies reacted with the proteins of the jellyfish structure in immunofluorescence microscopy if the cells were not permeabilized. These observations suggest that the structure is inside the cell. To confirm this, we treated the cell surface with sulfosuccinimidobiotin (Sulfo-NHS-biotin), which can modify the amino groups of side chains only when they are exposed on the exterior of a cell (36). Biotinization was not detected for the proteins of the jellyfish structure in blotting by avidin binding, although it was well detected for >10 surface proteins, including Gli349, MvspI, and so on (data not shown). This result shows that none of the jellyfish structure is exposed on the exterior of the cell.
Protein Components of the Jellyfish Structure.
We clarified the localizations of three component proteins of the jellyfish structure (Fig. 6 and SI Fig. 8). The MMOB1630 and MMOB4860 proteins were localized at the head, corresponding to the position of the bell observed by EM, and the MMOB1670 protein was localized at the cell neck, the position of the tentacles. These observations also suggest that the bell and tentacles of the jellyfish structure consist of different proteins (Fig. 2). Six of 10 ORFs identified for the jellyfish structure are coded tandemly with very short gaps on the genome (Fig. 5). On the other hand, six of seven ORFs in this locus code for the proteins of the jellyfish structure. These facts suggest that this locus may be responsible for the jellyfish structure.
MMOB4530 is annotated to an enzyme functioning in glycolysis, phosphoglycerate kinase, which transfers a phosphate group from 1,3-bisphosphoglycerate to ADP, yielding ATP and 3-phosphoglycerate. This protein may stay in the jellyfish structure and provide ATP to the motor for gliding. MMOB0150 is annotated to “xylose solute-binding protein” involved in the xylose transport system. This may suggest that the xylose transport system is localized at the head and/or neck through the association with the jellyfish structure.
Function of the F1-ATPase-Like Protein in Gliding.
The amino acid sequences of MMOB1660 and MMOB1670 show high similarity to the α- and β-subunits, respectively, of F1-ATPase, the catalytic subunit of proton pumps. For example, they have 38.8% identity in 420 residues with the F1-ATPase α-subunit and 48.0% identity in 427 residues with the F1-ATPase β-subunit of Bacillus subtilis. However, these proteins are unlikely to function in a proton pump, because the proton pump of B. subtilis requires an additional seven subunits (37), and the M. mobile genome has another locus containing the complete set of pump subunits.
At the base of the type III secretion system (TTSS) in Gram-negative bacteria, there is a protein subunit possessing high similarity to the F1-ATPase β-subunit, as represented by FliI of swimming flagella and InvC of the needle complex of Salmonella typhimurium (38, 39). These proteins function as a motor of the protein transporter. MMOB1660 and MMOB1670 may also function as the protein transporters for gliding proteins in M. mobile.
MMOB1670 is composed of 784 residues of amino acids, making it 311 and 314 residues longer than the proton pump β-subunits of B. subtilis and M. mobile, respectively. The comparison of these protein sequences showed that the amino acid sequence from the 1st to 299th residues in MMOB1670 is the extra region. Analyses by BLAST search showed that this extra region does not have similarities to any other proteins. This region is related to functions that are specific to M. mobile.
Azide, an inhibiter of proton pump ATPase, is known to react specifically to the catalytic subunits of the F1-ATPase β-subunit and its homologs (40). Thus, we examined the effect of this reagent on the gliding ghost to address the direct roles of MMOB1660 and MMOB1670 in gliding motility, but no effect was detected. This result may suggest that MMOB1660 and MMOB1670 are not directly involved in the gliding mechanism. P42 (MMOB1050), coded just downstream of three gliding proteins, was reported to have ATPase activity (21), and its characteristics are similar to those predicted from the behaviors of the gliding ghost (26). Therefore, this protein may function as the motor for gliding.
Comparison with the Architecture of M. pneumoniae.
M. pneumoniae also glides by means of a membrane protrusion, named “the attachment organelle,” formed at a cell pole (2–5, 46). The attachment organelle is known to have a cytoskeletal structure functioning as the scaffold for machinery of adherence and gliding (8, 31, 32, 41). This structure consists of three parts, a terminal button, rod, and wheel (bowl) from the distal end (42). The appearances of the terminal button and rod are reminiscent of the bell and tentacles of the jellyfish structure of M. mobile. To date, 11 protein components have been shown to be involved in the architecture of M. pneumoniae (2–7, 43, 46). Interestingly, none of them has similarity to the protein components of the jellyfish structure of M. mobile in terms of their amino acid sequences or, moreover, with any ORFs in the M. mobile genome (11). At the same time, no homologs of the proteins of the jellyfish structure can be found in the M. pneumoniae genome, except the phosphoglycerate kinase, MPN429, involved in glycolysis (44). The amino acid sequences of four gliding proteins, Gli123, Gli349, Gli521, or P42 also show no similarity to any of the ORFs of M. pneumoniae (20). These facts may suggest that the primary structures of gliding machinery are divergent between these two mycoplasma species. Some commonality may be found between the systems of the two species, if we could determine the three-dimensional structures of these proteins.
Conclusions
The mechanism supporting the strong force generated by the gliding machinery of M. mobile has been unexplained. In this study, we obtained critical information that led to our proposal of a structural model, the cytoskeletal jellyfish structure, which is distinct from any other bacterial cytoskeleton. It is hoped that this structure will shed light on our understanding of the bacterial cytoskeletons and their evolution.
Materials and Methods
Strains and Culture.
The wild-type and mutant strains of M. mobile strain 163K (ATCC 43663) were grown in Aluotto medium at 25°C (33). The mutant strains, m12, m13, and m9, mutated for the gli123, gli349, and gli521 genes, respectively, were reported previously (18–20, 33). The adhesive strain substituted for the gli521 gene as P476R has been reported (26). In the present study, this strain was used as the standard strain, when not otherwise mentioned, because the character of better binding is efficient for microscopy.
Electron Microscopy.
Mycoplasma cells suspended in the fresh medium at 10-fold density of the culture were put on a carbon-coated EM grid and incubated for 10 min at room temperature (RT). The medium was removed and replaced by Triton solution (0.01% to 1% Triton X-100, 1 mg/ml DNase, and 5 mM MgCl2 in PBS consisting of 75 mM sodium phosphate (pH 7.3), and 68 mM NaCl) for 1 min at RT. Triton solution was then removed, and the grid was stained for 1 min by 2% ammonium molybdate (vol/vol) and air-dried. When the jellyfish structures were compared among the strains, the cells in suspension were treated by the Triton solution containing 0.3% Triton X-100, put on EM grids, and then treated as described above. The samples were observed by an H-7000 transmission electron microscope (Hitachi) at 90 kV. Whole micrographs were digitized as 16-bit images by using DuoScan HiD (Agfa). Each particle image was picked up and averaged by EMAN, version 1.6 (http://ncmi.bcm.tmc.edu/∼stevel/EMAN/). Fourier transformation was done by Image J 1.37v (http://rsb.info.nih.gov/ij/).
Identifying Protein Components.
Mycoplasma cells were collected by centrifugation at 12,000 × g for 10 min at RT, washed twice with PBS/G (PBS containing 10 mM glucose), and resuspended by PBS/G to be 10-fold concentrated from the cell culture. The cells were filtered through a nitrocellulose filter (pore size, 0.45 μm) to disperse cells. The cells were treated by Triton solution (0.01–0.3% Triton X-100, 1 mg/ml DNase, and 5 mM MgCl2 in PBS) for 1 min at RT. The Triton-insoluble fraction was collected by centrifugation at 20,000 × g for 20 min at 4°C. The protein amounts were estimated by scanning and profiling the gels, as described (20). Focused protein bands were cut off and subjected to PMF by MALDI-TOF MS, as described (45). Mass spectra were measured in the reflectron mode of a MALDI-TOF mass spectrometer (AXIMA CFR Plus, Shimadzu/Kratos) operated in delayed extraction mode. Proteins were identified by an algorithm named Profound (http://prowl.rockefeller.edu).
Immunofluorescence and Immunoelectron Microscopy.
The polyclonal antibodies (antisera) were produced as described (17) by using the SDS/PAGE protein bands of mycoplasma cell fraction insoluble for Triton X-100. The specificity of antisera against the target proteins was confirmed by Western blotting. Mycoplasma cells of the standard strain suspended in a fresh medium were bound to cleaned glass and stained for the identified proteins, as described (6, 19). After chemical fixation by 3.0% paraformaldehyde and 0.1% glutaraldehyde for 30 min at RT, the cells were treated by 0.1% Triton X-100 in PBS for 1 min at RT if necessary. One-thousandfold and 100-fold dilutions were used for the antisera against the component proteins and Mvsps N and O, respectively.
For ImmunoGold EM, the cells bound to EM grids were treated by the Triton solution containing 0.3% Triton X-100 and fixed by 3% paraformaldehyde and 0.1% glutaraldehyde in PBS for 10 min at RT, and washed three times by PBS. The jellyfish structures on grids were treated by one 10-fold diluted antiserum against the component protein in PBS containing 2% BSA and washed five times by PBS. Then the jellyfish structures were treated by one 10-fold diluted gold-labeled secondary antibody (5 nm colloidal-gold-labeled goat antibody, Sigma) in PBS containing 2% BSA for 30 min at RT, washed five times, and then stained by 2% molybdate.
Supplementary Material
Acknowledgments
We thank Prof. Eisaku Katayama at Tokyo University for his helpful input. This work was supported by a Grant-in-Aid for Scientific Research (A), a Grant-in-Aid for Scientific Research on the Priority Areas “Structures of Biological Macromolecular Assemblies” and “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.M.), and by a grant from the Institution for Fermentation Osaka (to M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704280104/DC1.
References
- 1.Razin S, Yogev D, Naot Y. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krause DC, Balish MF. Mol Microbiol. 2004;51:917–924. doi: 10.1046/j.1365-2958.2003.03899.x. [DOI] [PubMed] [Google Scholar]
- 3.Miyata M. In: Mycoplasmas: Pathogenesis, Molecular Biology, and Emerging Strategies for Control. Blanchard A, Browning G, editors. Norfolk, U.K.: Horizon Bioscience; 2005. pp. 137–163. [Google Scholar]
- 4.Miyata M. Trends Microbiol. 2008 doi: 10.1016/j.tim.2007.11.002. in press. [DOI] [PubMed] [Google Scholar]
- 5.Miyata M, Ogaki H. J Mol Microbiol Biotechnol. 2006;11:256–264. doi: 10.1159/000094059. [DOI] [PubMed] [Google Scholar]
- 6.Seto S, Layh-Schmitt G, Kenri T, Miyata M. J Bacteriol. 2001;183:1621–1630. doi: 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seto S, Miyata M. J Bacteriol. 2003;185:1082–1091. doi: 10.1128/JB.185.3.1082-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu T, Miyata M. Curr Microbiol. 2002;44:431–434. doi: 10.1007/s00284-001-0014-8. [DOI] [PubMed] [Google Scholar]
- 9.Bredt W. Pathol Microbiol. 1968;32:321–326. doi: 10.1159/000162074. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhoff H. In: Mycoplasmas—Molecular Biology and Pathogenesis. Maniloff J, McElhaney RN, Finch LR, Baseman JB, editors. Washington, DC: Am Soc Microbiol; 1992. pp. 289–306. [Google Scholar]
- 11.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, et al. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiratsuka Y, Miyata M, Tada T, Uyeda TQP. Proc Natl Acad Sci USA. 2006;103:13618–13623. doi: 10.1073/pnas.0604122103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiratsuka Y, Miyata M, Uyeda TQP. Biochem Biophys Res Commun. 2005;331:318–324. doi: 10.1016/j.bbrc.2005.03.168. [DOI] [PubMed] [Google Scholar]
- 14.Miyata M, Ryu WS, Berg HC. J Bacteriol. 2002;184:1827–1831. doi: 10.1128/JB.184.7.1827-1831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyata M, Uenoyama A. FEMS Microbiol Lett. 2002;215:285–289. doi: 10.1111/j.1574-6968.2002.tb11404.x. [DOI] [PubMed] [Google Scholar]
- 16.Rosengarten R, Kirchhoff H. J Bacteriol. 1987;169:1891–1898. doi: 10.1128/jb.169.5.1891-1898.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusumoto A, Seto S, Jaffe JD, Miyata M. Microbiology. 2004;150:4001–4008. doi: 10.1099/mic.0.27436-0. [DOI] [PubMed] [Google Scholar]
- 18.Seto S, Uenoyama A, Miyata M. J Bacteriol. 2005;187:3502–3510. doi: 10.1128/JB.187.10.3502-3510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uenoyama A, Kusumoto A, Miyata M. J Bacteriol. 2004;186:1537–1545. doi: 10.1128/JB.186.5.1537-1545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uenoyama A, Miyata M. J Bacteriol. 2005;187:5578–5584. doi: 10.1128/JB.187.16.5578-5584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani N, Miyata M. Biochem J. 2007;403:71–77. doi: 10.1042/BJ20061439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adan-Kubo J, Uenoyama A, Arata T, Miyata M. J Bacteriol. 2006;188:2821–2828. doi: 10.1128/JB.188.8.2821-2828.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyata M, Petersen J. J Bacteriol. 2004;186:4382–4386. doi: 10.1128/JB.186.13.4382-4386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe JD, Miyata M, Berg HC. J Bacteriol. 2004;186:4254–4261. doi: 10.1128/JB.186.13.4254-4261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai R, Miyata M. J Bacteriol. 2006;188:6469–6475. doi: 10.1128/JB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uenoyama A, Miyata M. Proc Natl Acad Sci USA. 2005;102:12754–12758. doi: 10.1073/pnas.0506114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charon NW. Proc Natl Acad Sci USA. 2005;102:13713–13714. doi: 10.1073/pnas.0506508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carballido-Lopez R, Errington J. Trends Cell Biol. 2003;13:577–583. doi: 10.1016/j.tcb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Moller-Jensen J, Lowe J. Curr Opin Cell Biol. 2005;17:75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Bredt W. In: The Mycoplasmas. Barile MF, Razin S, Tully JG, Whitcomb RF, editors. Vol 1. New York: Academic; 1979. pp. 141–145. [Google Scholar]
- 31.Henderson GP, Jensen GJ. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- 32.Seybert A, Herrmann R, Frangakis AS. J Struct Biol. 2006;156:342–354. doi: 10.1016/j.jsb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Miyata M, Yamamoto H, Shimizu T, Uenoyama A, Citti C, Rosengarten R. Microbiology. 2000;146:1311–1320. doi: 10.1099/00221287-146-6-1311. [DOI] [PubMed] [Google Scholar]
- 34.Deckers-Hebestreit G, Altendorf K. Annu Rev Microbiol. 1996;50:791–824. doi: 10.1146/annurev.micro.50.1.791. [DOI] [PubMed] [Google Scholar]
- 35.Kirchhoff H, Rosengarten R, Lotz W, Fischer M, Lopatta D. Israel J Med Sci. 1984;20:848–853. [PubMed] [Google Scholar]
- 36.Dorsey CW, Tolmasky ME, Crosa JH, Actis LA. Microbiology. 2003;149:1227–1238. doi: 10.1099/mic.0.26204-0. [DOI] [PubMed] [Google Scholar]
- 37.Santana M, Ionescu MS, Vertes A, Longin R, Kunst F, Danchin A, Glaser P. J Bacteriol. 1994;176:6802–6811. doi: 10.1128/jb.176.22.6802-6811.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imada K, Minamino T, Tahara A, Namba K. Proc Natl Acad Sci USA. 2007;104:485–490. doi: 10.1073/pnas.0608090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akeda Y, Galan JE. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 40.Vasilyeva EA, Minkov IB, Fitin AF, Vinogradov AD. Biochem J. 1982;202:15–23. doi: 10.1042/bj2020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biberfeld G, Biberfeld P. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hegermann J, Herrmann R, Mayer F. Naturwissenschaften. 2002;89:453–458. doi: 10.1007/s00114-002-0359-2. [DOI] [PubMed] [Google Scholar]
- 43.Kenri T, Seto S, Horino A, Sasaki Y, Sasaki T, Miyata M. J Bacteriol. 2004;186:6944–6955. doi: 10.1128/JB.186.20.6944-6955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sanchez-Pulido L, Snel B, Suyama M, et al. Nucleic Acids Res. 2000;28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellman U, Wernstedt C, Gonez J, Heldin CH. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 46.Miyata M. In: Cell Motility. Lenz P, editor. Berlin: Springer; in press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.