Figure 1. An RNA Element Derived from a Replication-Dependent Histone mRNA Stimulates 3′-end Formation.
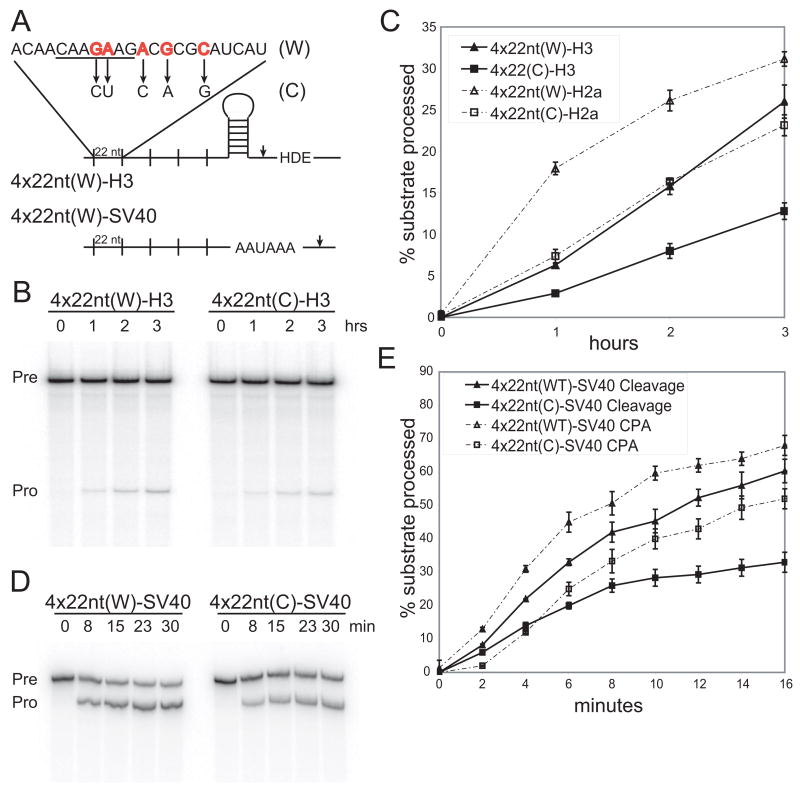
(A) The U7-snRNP (derived from mouse histone H3 mRNA) and CPA (SV40 late) substrates are schematized with sites of cleavage indicated by arrowheads. The 22-nt RNA element (W) and a control element (C), with changed nucleotides indicated by arrows, have been described (Huang and Steitz, 2001). The 7-nt conserved motif is underlined.
(B and C) Time courses of U7-snRNP-dependent cleavage of two U7-snRNP substrates (H3 and H2a) containing four copies of the 22-nt W element or the C element were performed in HeLa nuclear extract (Kolev and Steitz, 2005). The H3 substrate (Pre→Pro) is shown in B; processing of both substrates was quantitated (error bars represent standard deviations; panel C) as the ratio of the 5′ product to total RNA. We verified that the substrates are cleaved in a U7-snRNP-dependent manner (data not shown; Bond et al., 1991; Huang et al., 2003).
(D and E) Processing time courses of the SV40 late polyadenylation substrate (Pre) with four copies of either the 22-nt W element or the C element were performed in HeLa nuclear extract (Ryan et al., 2004); note the shorter time required for comparable extent of cleavage in E versus C). In D, cordycepin was added to inhibit polyadenylation of the cleavage product (Pro). E shows quantitations, as in panel C, for both the uncoupled and coupled CPA reactions.