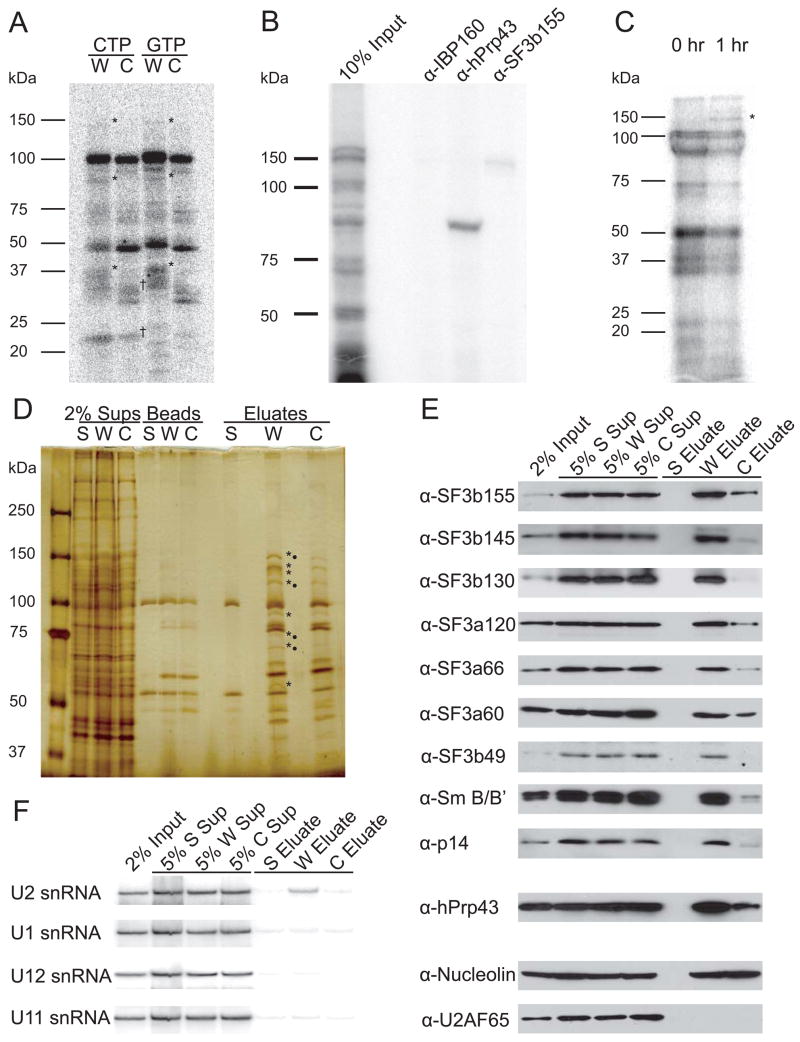
Figure 2. U2 snRNP Binds the 22-nt RNA Element In Vitro and In Vivo.
(A)RNAs containing four copies of the 22-nt (W) or control (C) element were UV cross-linked under CPA conditions (Ryan et al., 2004). RNAs were synthesized in the presence of 32P-labeled CTP or GTP as indicated, and cross-linked proteins were separated by 5–15% gradient SDS-PAGE. The 22-nt RNA element (W) cross-linked specifically to three proteins (*) compared to the control RNA (C). SRp20 and 9G8, previously characterized as cross-linking to the RNA element (Huang and Steitz, 2001), are detected for both RNAs labeled at C-residues (†).
(B) A CTP-labeled RNA containing four copies of the 22-nt element (W) was incubated and UV-irradiated as in A; immunoprecipitations were performed with the indicated antibodies followed by 5–15% gradient SDS-PAGE.
(C) CTP-labeled RNA containing the complete H2a 101-nt RNA element 5′ to the H3-derived U7-snRNP substrate was injected into GVs and UV-irradiated after 0 or 1 hr; cross-linked proteins were separated by 12% SDS-PAGE. Proteins visible at zero time most likely represent a pre-processing heterogeneous RNP, as described for in vitro CPA (Skolnik-David et al., 1987) and serve as a control. A specific 150 kDa cross-linked protein was observed (*), but could not be identified by α-SF3b155 immunoprecipitation because the signal was too weak.
(D) Affinity purification was performed using the biotinylated 22-nt RNA element (W, in four copies), the mutated control (C, in four copies), or streptavidin beads (S) alone. Supernatants (Sups), proteins retained on the beads after RNase elution (Beads), and the eluted proteins (Eluates) were separated by 5–15% gradient SDS-PAGE. Proteins enriched in the purification with W relative to C are indicated (*), as are proteins identified by mass spectrometric analysis (•).
(E) Western blots were performed on affinity-purified samples to confirm and extend the mass spectrometry results. U2B″ and U2A′ were also enriched in W versus C (data not shown).
(F) Northern blotting identified U2 snRNA in the eluate from the W affinity purification.