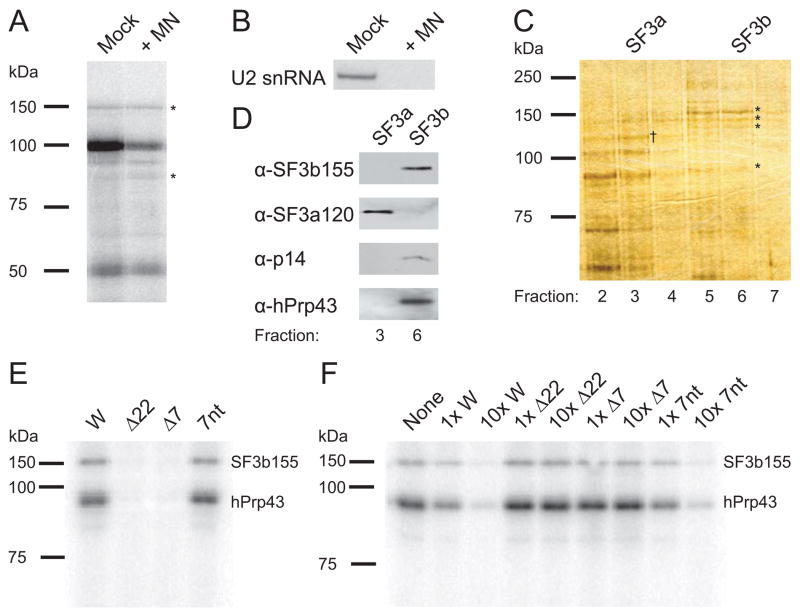
Figure 3. Isolated SF3b/hPrp43 Requires the 7-nt RNA Motif for Binding.
(A) Nuclear extract digested with micrococcal nuclease (MN) or mock treated was incubated with [α-32P]-CTP-labeled RNA containing four copies of the 22-nt RNA element and UV-irradiated. The profile of cross-linked proteins separated by 8% SDS-PAGE indicated comparable cross-linking of the 150 and 93 kDa proteins (*).
(B) A Northern blot was performed to confirm destruction of U2 snRNA by MN-treatment.
(C) Glycerol gradient fractions 2–7 (top-bottom) from the SF3b/hPrp43 purification were analyzed by 8% SDS-PAGE and silver stained. SF3b155, SF3b145, SF3b130, and hPrp43 are visible in fractions 5–7 (*), whereas SF3a components peak in fraction 3 (SF3a120 is indicated, †). A similarly sized band to hPrp43 is seen in fractions 2–4, but must be a different protein (see D below).
(D) Fractions containing SF3b (fraction 6) and SF3a (fraction 3) were compared by Western blotting for SF3b155, SF3a120, p14, and hPrp43. Fraction 3 does not contain detectable levels of hPrp43.
(E) Purified SF3b/hPrp43 was tested for cross-linking in vitro to the H2a 101-nt element (W) and to mutant RNAs with either the 22-nt RNA element (Δ22) or conserved 7-nt motif (Δ7) deleted or a 37-nt RNA containing the 7-nt motif flanked by different sequences (7nt; see Supplemental Data). Three independent experiments showed >10-fold stronger cross-linking to W than to either mutant.
(F) Purified SF3b/hPrp43 was cross-linked in vitro to the radiolabeled H2a 101-nt RNA element in the presence of the indicated unlabeled competitor RNAs at low (1x) and high (10x) concentrations. Comparable results were obtained in three independent experiments. The band at ~80 kDa is variable, probably a breakdown product.