Abstract
The precise location of the tRNA processing ribonucleoprotein ribonuclease P (RNase P) and the mechanism of its intranuclear distribution have not been completely delineated. We show that three protein subunits of human RNase P (Rpp), Rpp14, Rpp29 and Rpp38, are found in the nucleolus and that each can localize a reporter protein to nucleoli of cells in tissue culture. In contrast to Rpp38, which is uniformly distributed in nucleoli, Rpp14 and Rpp29 are confined to the dense fibrillar component. Rpp29 and Rpp38 possess functional, yet distinct domains required for subnucleolar localization. The subunit Rpp14 lacks such a domain and appears to be dependent on a piggyback process to reach the nucleolus. Biochemical analysis suggests that catalytically active RNase P exists in the nucleolus. We also provide evidence that Rpp29 and Rpp38 reside in coiled bodies, organelles that are implicated in the biogenesis of several other small nuclear ribonucleoproteins required for processing of precursor mRNA. Because some protein subunits of RNase P are shared by the ribosomal RNA processing ribonucleoprotein RNase MRP, these two evolutionary related holoenzymes may share common intranuclear localization and assembly pathways to coordinate the processing of tRNA and rRNA precursors.
Keywords: coiled body, nucleolus, RNase mitochondrial RNA processing, ribonuclease P, tRNA
Many processes of fundamental importance to the usage of genetic information in eukaryotes take place, or have their catalytic components assembled, in the nucleolus (for review see Melese and Xue 1995; Shaw and Jordan 1995; Lamond and Earnshaw 1998; Pederson 1998). Events related to processes critical for the cell cycle, life span, and apoptosis also occur in the nucleoli in some eukaryotes (for review see Guarente 1997; Bachant and Elledge 1999). Ribonucleoprotein complexes with catalytic roles in RNA processing and modification are major constituents of nucleoli (Pederson et al., 1998; Yu et al. 1999). These ribonucleoproteins have to find their way to the nucleolus or even to certain compartments within these dynamic structures and to the nearby coiled bodies. Specific sequences in certain proteins have already been identified that function, alone or in concert with sequences in other proteins or nucleic acids, to achieve nucleolar localization. However, to date there is no apparent consensus sequence in proteins that determines nucleolar localization, probably because a variety of different protein–protein and protein–nucleic acid interactions are used in the targeting process. In this report, we describe distinct subnucleolar localization domains found in two protein subunits of the human tRNA processing ribonucleoprotein ribonuclease P (RNase P).1
The precise locations of RNase P in eukaryotic cells have not been completely delineated (Matera et al. 1995; Lee et al. 1996; Jacobson et al. 1997; Pederson 1998; for review see Wolin and Matera 1999), although it is well established that processing of the 5′ termini of some precursor tRNAs is a nuclear (Melton and Cortese 1979) or nucleolar event (Bertrand et al. 1998). The RNA subunit of human RNase P has been identified in the cytoplasm, nucleoplasm, the perinucleolar compartment, as well as the nucleolus (Matera et al. 1995; Lee et al. 1996; Jacobson et al. 1997), but the majority is nucleoplasmic.
Several proteins have been characterized as subunits of human RNase P (Lygerou et al. 1996; Eder et al. 1997; Jarrous et al. 1998, Jarrous et al. 1999). Extensive sharing of protein components of the yeast nuclear RNase P and the rRNA processing enzyme RNase MRP, have now been established by genetic and some biochemical means (Chamberlain et al. 1998). Although the protein composition of human RNase MRP remains to be verified by extensive biochemical purification analysis, several RNase P protein subunits are shared by RNase MRP (Lygerou et al. 1996; Eder et al. 1997; Jarrous et al. 1999; Pluk et al. 1999). The specific functions of these protein subunits in RNase P and RNase MRP assembly and intracellular localization, however, remain unknown.
We show here that several protein subunits of human RNase P are primarily localized in the nucleolus of mammalian cells, as determined by confocal immunofluorescence microscopy. Two RNase P protein(Rpp) subunits, Rpp14 and Rpp29, are localized in the dense fibrillar component, whereas the other subunit, Rpp38, is more uniformly distributed in the nucleolus. Rpp29 and Rpp38 possess functional sequences required for nucleolar localization. Rpp14 appears to enter the nucleolus through a piggyback process. Rpp29 and Rpp38 are also found in coiled bodies, nucleoplasmic structures that participate in the transport and sorting of several small nuclear and nucleolar ribonucleoprotein components involved in the processing of mRNA and rRNA precursors as well as in the 3′ end formation of histone mRNA precursors (Gall et al. 1995; Lamond and Earnshaw 1998).
Materials and Methods
Cell Cultures and Transfection
Mouse Swiss 3T3 fibroblasts, HeLa cells, and human embryonic kidney 293 cells were grown in high glucose DME (Life Technologies, Inc.) supplemented with L-glutamine, 25 mM Hepes buffer, pyridoxine hydrochloride, 10% FBS, streptomycin (100 μg/ml), and penicillin (100 U/ml). Cells were incubated in 5% CO2 at 37°C in 75-cm2 flasks. For transient transfections, 1–5 × 105 cells grown in 60 × 15 mm style petri dishes containing glass coverslips were transfected with plasmid DNA (2–5 μg) using the SuperFect reagent (Qiagen) following the manufacturer's instructions. For stable transfection, 1–2 × 106 HEK 293 cells were transfected with 2 μg of pEGFP-Rpp38, split 24 h after transfection, and then grown in a selective medium supplemented with 0.5 mg/ml geneticin (G418; Life Technologies, Inc.). Mixtures of G418-resistant cell populations obtained were massively grown for RNase P purification.
Gene Constructs
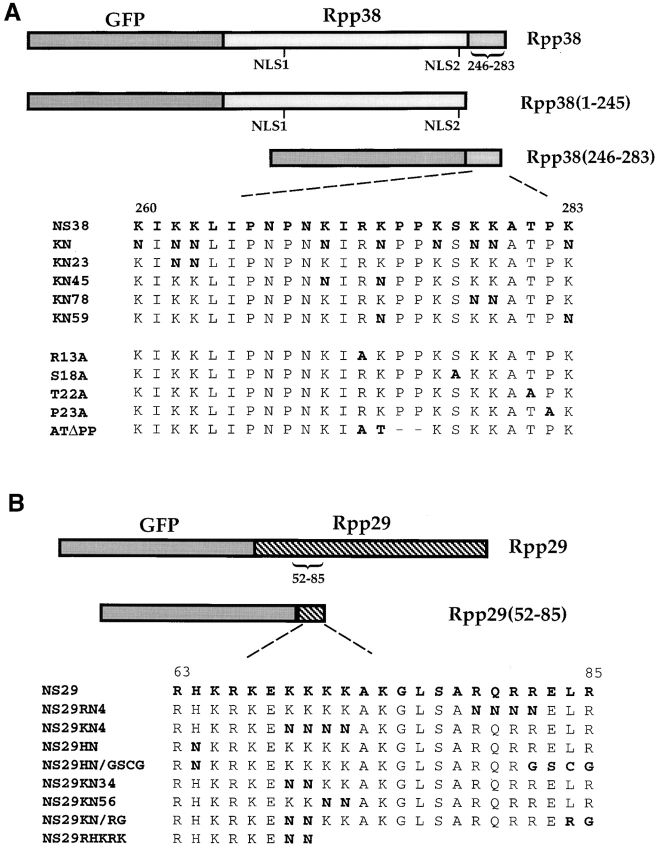
A PstI-NotI Rpp38 cDNA (Eder et al. 1997) fragment subcloned in pBluescript was released by PstI and ApaI (located in the multiple cloning site) and subcloned in-frame in PstI-ApaI digested pEGFP-C1 (CLONTECH Laboratories) to generate pEGFP-Rpp38. pEGFP-Rpp38(246-283) was generated by cleaving pEGFP-Rpp38 with HindIII, deleting the first 245 amino acids of Rpp38, and then the plasmid was self-ligated in the presence of a short HindIII DNA adaptor to keep the carboxy terminal 37 amino acids of Rpp38 (positions 246–283) in-frame with GFP. pEGFP-Rpp38(1-245) was constructed by cleaving a PstI-HindIII Rpp38 cDNA (Eder et al. 1997) subcloned in pBluescript with PstI and ApaI and subcloned in-frame in pEGFP-C1 first cleaved with PstI and ApaI.
pEGFP-Rpp38(260-283) was generated by subcloning a PstI-ApaI deoxyoligonucleotide that codes for the last 24 amino acids of Rpp38 in pEGFP-C1 digested with PstI and ApaI. pNS38KN was constructed as pEGFP-Rpp38(260-283) with all the nine lysine residues in the carboxy terminal 24–amino acid sequence were substituted with asparagines. pNS38KN23, pNS38KN45, pNS38KN78, and pNS38KN59 were constructed as pEGFP-Rpp38(260-283) but with two lysine substitutions; numbers represent the substituted lysines (see Fig. 1 A). Constructs with a single substitution of arginine (pNS38R13A), serine (pNS38S18A), threonine (pNS38T22A), or proline (pNS38P23A) to alanine (see positions in Fig. 1 A) were also prepared as described for pEGFP-Rpp38(260-283). pNS38ATΔPP was obtained during the construction of pNS38R13A in which we found after sequencing that the arginine and lysine were substituted accidentally by alanine and threonine, respectively, whereas the consecutive proline residues were deleted, keeping the remaining amino acids in-frame with the upstream GFP.
Figure 1.
Schematic representation of gene constructs. (A) Rpp38 or portions of this protein, Rpp38(1-245) or Rpp38(246-283), was fused in-frame to GFP in pEGFP-C1 expression vector (see Materials and Methods). Numbers indicate positions of residues in the 283–amino acid Rpp38 polypeptide. NLS1 and NLS2 indicate putative NLSs. NS38 represents the amino acid sequence from position 260–283 of Rpp38. Substitution mutations of the nine lysines to asparagines in NS38 sequence, and their numbers, are presented in bold letters. Single substitution mutants of arginine (R13A), serine (S18A), threonine (T22A), and proline (P23A) to alanine are also shown. ATΔPP has substitution of arginine and lysine with alanine and threonine, respectively, and the two consecutive proline residues were deleted from NS38. (B) Full-length Rpp29 or the amino acids encompassing positions 52–85 or 63–85 of Rpp29 were fused in-frame with GFP in pEGFP-C1. The amino acid sequence of Rpp29 from position 63–85, designated NS29, is indicated. Several mutants of NS29 with single and multiple substitutions or deletions are shown. All gene fusion constructs were transcribed from a cytomegalovirus promoter-enhancer located upstream of GFP in the expression plasmid.
Two primers, one encompassing the translation initiation codon and the other one spanning the translation stop codon of Rpp29, were used to amplify the entire Rpp29 cDNA (Jarrous et al. 1999). The PCR product was digested with EcoRI, located in the designed primer sequences, and subcloned in-frame in pEGFP-C1 first cleaved with EcoRI. pEGFP-Rpp29(52-85) was constructed by digestion of a PCR DNA product, containing the sequence that codes for amino acids 52–85 of Rpp29 (see Fig. 1 B), with Hind III and BamHI located in the designed primers, and followed by subcloning in pEGFP-C1 cleaved first with the same two restriction enzymes. pEGFP-Rpp29(63-85) was generated by subcloning a BamHI-HindIII deoxyoligonucleotide that codes for the 23 amino acids encompassing positions 63–85 of Rpp29 in pEGFP-C1 digested with BamHI and HindIII. pNS29RN4 and pNS29KN4 were constructed as pEGFP-Rpp29(63-85), but the RQRR or KKKK residues were substituted with four asparagines, respectively. pEGFP-Rpp14 was generated by subcloning a XhoI-HindIII–digested PCR DNA product containing the entire Rpp14 open reading frame (Jarrous et al. 1999) into pEGFP-C1 cleaved first with XhoI and HindIII.
pPK-Rpp38 was generated by inserting a KpnI-digested PCR Rpp38 cDNA containing the entire open reading frame into the KpnI unique site of the myc-tagged chicken pyruvate kinase in pcDNA3-PK plasmid (Siomi and Dreyfuss 1995), provided to us by Dr. Gideon Dreyfuss (University of Pennsylvania, Philadelphia, PA). All DNA constructs described above were verified by sequencing to ensure in-frame subcloning of the desired inserts with the reporter gene.
Indirect Immunofluorescence
Cells (20% confluent) were grown overnight on coverslips (22 × 22 mm) before fixation with 2% paraformaldehyde (Electron Microscopy Sciences) diluted in 1× PBS for 30 min. Cells were treated with 0.5% Triton X-100 for 5–30 min, washed twice with 1× PBS (0.5 liter each), and then blocked with 3% BSA/PBS for 20 min. Rabbit polyclonal antibodies against Rpp subunits (Jarrous et al. 1999), p80-coilin (Andrade et al. 1993), or Nopp140 peptide (Meier and Blobel 1992), diluted 1:50–400 in 3% BSA/PBS, were added to the fixed cells for 1 h, and then washed twice with PBS before incubation for 20 min with 1:50 diluted secondary antibody, Alexa™ 568 goat anti–rabbit IgG antibody conjugate (Molecular Probes Inc.). When the monoclonal antifibrillarin and anti-myc (9E10) mouse IgG antibodies or polyclonal anti-B23 goat IgG antibodies (Santa Cruz Biotechnology Inc., CA) were included, Alexa™ 488 goat anti–mouse IgG antibody or Alexa™ 594 donkey anti–goat IgG antibody conjugates (Molecular Probes Inc.) were used. Cells were washed twice with PBS and mounted on glass slides using boat sealer (Ernest Fullam).
Microscopy and Imaging
Confocal fluorescence microscopy of living or fixed cells was performed at 22°C (± 2°C) using a Bio-Rad MRC-1024 laser scanner mounted on a 2FL reflector slider on a Zeiss Axiovert equipped with differential interference contrast (DIC) optics (PlanApo 100× 1.4 NA oil immersion objective; Carl Zeiss). Fluorescent images were acquired by using Texas red and FITC filters, and then processed using LaserSharp software (Bio-Rad Laboratories). Bleedthrough was completely eliminated between fluorophore channels in colocalization studies. Nuclei of living cells were also visualized by DNA staining with 4′,6-diamidino-2-phenylindole. Digital processing and color adjustment of images were done using MetaMorph Image acquisition and processing software (Universal Imaging Corp.) and Adobe Photoshop (Adobe Systems, Inc.).
Purification and Analysis of Human RNase P
RNase P from G418-resistant 293 HEK cells that constitutively express GFP-Rpp38 fusion protein was purified as previously described (Eder et al. 1997). In brief, 109 cells were pelleted, disrupted, and the cell homogenate was centrifuged at 7,000 rpm followed by another centrifugation at 42,000 rpm in a Beckman Ti50 rotor to obtain S100 crude extract. This S100 extract was loaded on a DEAE-Sepharose anion exchange chromatography column and RNase P was eluted from the column using a 100–500-mM KCl gradient. The flowthrough, wash, and the eluted fractions were assayed for RNase P activity, and then kept in 25% glycerol in −20°C for further analysis. Cleavage of the 5′ leader of the yeast suppressor precursor tRNASer (SupS1) by human RNase P was performed as described (Jarrous et al. 1998). For Western blot analysis, DEAE fractions were separated on 12% SDS–polyacrylamide gel, electrotransferred to a nitrocellulose filter, and immunoblotted with 1:3,000 diluted polyclonal anti-GFP antibodies (CLONTECH Laboratories) or with 1:100 dilution of affinity-purified polyclonal anti-Rpp38 rabbit antibodies (Jarrous et al. 1998). Peroxidase-labeled goat anti–rabbit IgG antibodies (Vector Labs, Inc.) were used at 1:5,000 dilution as secondary antibodies. Blots were washed and bands were visualized using the ECL plus kit (Amersham), following the manufacturer's instructions.
Results
Rpp38 Localizes a Green Fluorescent Protein (GFP) to the Cell Nucleolus and Coiled Bodies
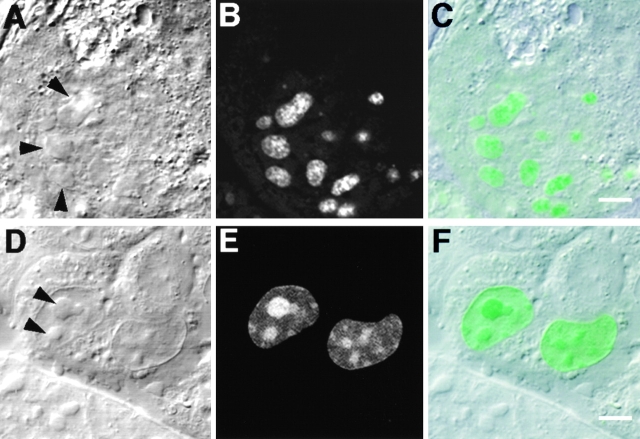
Mouse Swiss 3T3 fibroblasts were transiently transfected with pEGFP-Rpp38, a derivative of the expression vector pEGFP-C1, which contains the Rpp38 open reading frame fused in-frame to the carboxy terminus of a GFP (Fig. 1 A). Expression of the GFP-Rpp38 fusion protein in transfected cells was monitored by confocal fluorescence microscopy (see Materials and Methods). 48 h after transfection, the fluorescence signal of GFP-Rpp38 was seen in the nucleoplasm but was most visible in the nucleoli (Fig. 2A and Fig. B, and see below). Only background fluorescence was observed in the cytoplasm. By contrast, GFP alone was distributed diffusely throughout the cytoplasm and the nucleoplasm, but was completely excluded from nucleoli (Fig. 2C and Fig. D). Since GFP-Rpp38 was not seen in the cytoplasm, as was GFP alone, this fusion protein must have been retained in the nucleoplasm and nucleoli.
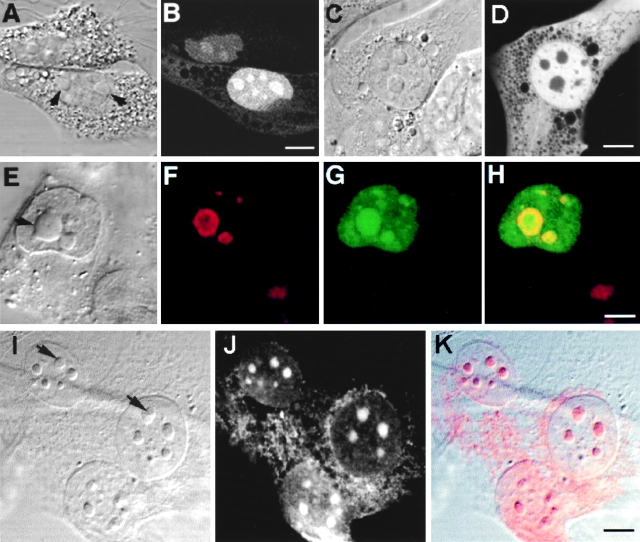
Figure 2.
Intracellular localization of Rpp38. Swiss 3T3 fibroblasts were transiently transfected (see Materials and Methods) with pEGFP-Rpp38 (A and B) or pEGFP-C1 (C and D). 48 h after transfection, living cells were examined with a confocal microscope. Colocalization of GFP-Rpp38 in the nucleolus with the protein B23 in transfected HeLa cells is demonstrated by indirect immunofluorescence analysis (E–H). (I–K) Immunofluorescence of endogenous Rpp38 protein in untransfected 3T3 fibroblasts using anti-Rpp38 antibodies showing uniformly stained nucleoli. Images of DIC (A, C, E, and I), GFP (B, D, and G), B23 (F), and anti-Rpp38 (J) are shown. H is an overlay of F and G, and K is an overlay of I and J. No specific signal above autofluorescence was seen when control sera of rabbits were tested (not shown). Arrows point to nucleoli. Bars: B, D, and K, 5 μm; H, 2.5 μm.
Localization of GFP-Rpp38 in the nucleolus of transfected 3T3 fibroblasts was verified by the colocalization of this fusion protein with the nucleolar protein B23 (Biggiogera et al. 1990) using indirect immunofluorescence analysis (Fig. 2, E–H). B23 is a nuclear localization sequence (NLS)–binding phosphoprotein that is found in the dense fibrillar component and the granular component of the nucleolus (Biggiogera et al. 1990). GFP-Rpp38 is more uniformly distributed than B23 in the nucleolar compartments (Fig. 2, E–H).
That endogenous Rpp38 in 3T3 fibroblasts is also a nucleolar protein was confirmed by using affinity-purified, polyclonal anti-Rpp38 antibodies (Jarrous et al. 1998) in indirect immunofluorescence analysis (Fig. 2, I–K). As with GFP-Rpp38 (Fig. 2 B), endogenous Rpp38 was uniformly distributed in the nucleolus. A weak signal around the nucleus that is typical of mitochondrial staining was also observed (Fig. 2, I–K), but further work is required to confirm localization of Rpp38 in these cytoplasmic organelles.
We also tested the ability of Rpp38 to target another reporter protein, the cytoplasmic chicken pyruvate kinase (Siomi and Dreyfuss 1995). A fusion protein of Rpp38 with a myc-tagged pyruvate kinase accumulated in the nucleoplasm of transfected 3T3 cells and a weak signal was seen in nucleoli (data not shown). This may suggest that this fusion protein is too large (∼100 kD) to be efficiently translocated and/or retained in the nucleolus, when compared with GFP-Rpp38. Therefore, GFP was used as the reporter protein throughout this study.
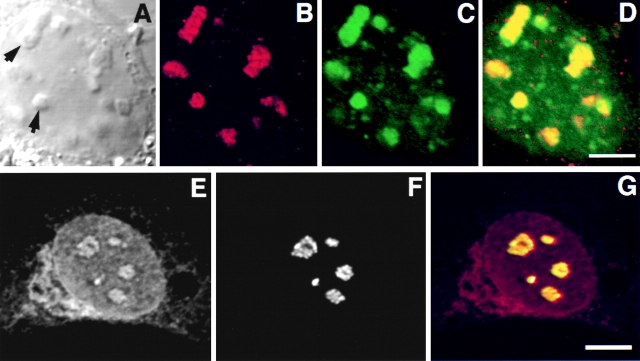
In transfected HeLa cells, GFP-Rpp38 compartmentalized in nucleoli as well as in discrete, intranuclear organelles immunostained with an antibody against p80-coilin (Fig. 3, A–D). These organelles represent coiled bodies as defined by the presence of p80-coilin (Andrade et al. 1993; Bauer et al. 1994). Diffuse immunostaining of p80-coilin was seen in the nucleoplasm and the nucleolus as well (Fig. 3, A–D; Lamond and Earnshaw 1998). The site of Rpp38 was further identified by the colocalization of GFP-Rpp38 with the nucleolar shuttling protein Nopp140 (Meier and Blobel 1992; Isaac et al. 1998) in nucleoli and coiled bodies (Fig. 3, E–H). Nopp140 is confined to the dense fibrillar component of the nucleolus (Meier and Blobel 1992). Clearly, GFP-Rpp38 is more widely distributed in nucleoli than Nopp140. Similar results regarding coiled bodies were obtained with transfected 3T3 fibroblasts (data not shown). All these findings, taken together, demonstrate that the Rpp38 subunit of RNase P is localized in the nucleolus and in coiled bodies of cultured mammalian cells.
Figure 3.
GFP-Rpp38 is localized in the nucleoli and coiled bodies. HeLa cells were transfected for 48 h with pEGFP-Rpp38, and then were examined for the presence of p80-coilin (A–D) or Nopp140 (E–H) in indirect immunofluorescence analysis. DIC (A and E), p80-coilin (B, red), Nopp140 (F, red), GFP-Rpp38 (C and G, green), and the overlays of B over C and F over G are shown in D and H, respectively. Intense yellow color is seen in the nucleoli and coiled bodies indicated by arrows. Overlays were acquired at the same confocal plane using Texas red (red) and FITC (green) filters. Bleedthrough between the two channels was completely eliminated. 3–6 coiled bodies were usually seen in HeLa cells. Inserts shown in A–D represent higher magnification of coilin-immunostained coiled bodies in proximity to a nucleolus of HeLa cells expressing GFP-Rpp38. All images were acquired during the same experimental observation. Bars: D and H, 2 μm; insert, 0.5 μm.
The Domain for Nucleolar Localization Resides in the Carboxy Terminus of Rpp38
Examination of the amino acid sequence of Rpp38 shows that it may possess three NLSs, located at positions 63–66, 241–244, and 262–281 (Fig. 1 A; Jarrous et al. 1998). Two DNA constructs, pEGFP-Rpp38(1-245), which contains amino acids 1–245 of Rpp38 and pEGFP-Rpp38(246-283), which possesses the remaining 37 amino acids of the polypeptide (see Fig. 1 A), were separately transfected into 3T3 fibroblasts and the localization of these truncated fusion proteins was determined. GFP-Rpp38(1-245) was concentrated in the nucleoplasm but not in the nucleoli (Fig. 4, A–C), an observation that was also confirmed in a double label experiment in transfected cells immunostained for the nucleolar B23 protein (Fig. 4, G–J). By contrast, GFP-Rpp38(246-283) was targeted to the nucleoli and nucleoplasm of cells with no significant signal seen in the cytoplasm (Fig. 4, D–F and K–N, and Table ). Similar nuclear and nucleolar localization patterns were obtained in 293 HEK cells transfected with the two constructs described above (not shown). Thus, the sequence required for nucleolar localization of Rpp38 exists between positions 246–283.
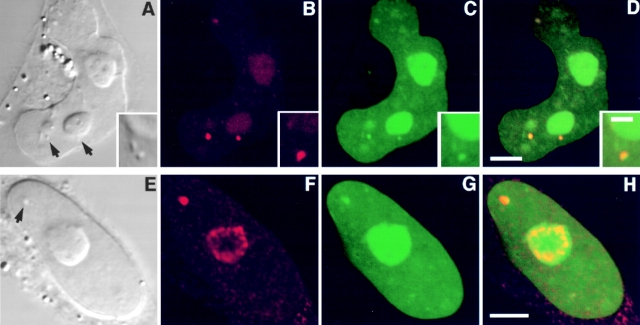
Figure 4.
A nucleolar localization signal exists in the carboxy terminus of Rpp38. Swiss 3T3 fibroblasts were transfected with pEGFP-Rpp38(1-245) (A–C and G–J) or with pEGFP-Rpp38(246-283) (D–F and K–N). Construct maps are shown in Fig. 1 A. 48 h after transfection, living cells were examined (A–F) using confocal microscopy, or first fixed and subjected to indirect immunofluorescence using antibody against nucleolar protein B23 (G–J and K–N). DIC (A, D, G, and K), GFP (B, E, I, and M, green), B23 (H and L, red), and overlays (C, F, J, and N) are shown. Arrows point to nucleoli. Bars: C and F, 5 μm; J and N, 2.5 μm.
Table 1.
Relative Intensities of Fluorescence in Three Different Compartments of 3T3 Fibroblasts Transfected with the GFP Fusion Constructs
| Construct | Cellular localization | ||
|---|---|---|---|
| Cytoplasm | Nucleoplasm | Nucleoli | |
| pEGFP | ++ | ++ | − |
| pEGFP-Rpp38 | −+ | ++ | +++ |
| pEGFP-Rpp38(1-245) | −+ | +++ | − |
| pEGFP-Rpp38(246-283) | −+ | ++ | +++ |
| pNS38 | −+ | ++ | +++ |
| pNS38KN | +++ | ++ | − |
| pNS38KN23 | ++ | ++ | ++ |
| pNS38KN45 | ++ | ++ | ++ |
| pNS38KN78 | + | +++ | − |
| pNS38KN59 | + | ++ | ++ |
| pNS38R13A | + | ++ | ++ |
| pNS38S18A | −+ | ++ | ++ |
| pNS38T22A | −+ | ++ | ++ |
| pNS38P23A | −+ | + | +++ |
| pNS38ATDPP | + | ++ | ++ |
| pEGFP-Rpp29 | − | + | +++ |
| pEGFP-Rpp29(52-85) | −+ | + | +++ |
| pEGFP-Rpp29(63-85) | −+ | + | +++ |
| pNS29RN4 | −+ | + | +++ |
| pNS29KN4 | ++ | ++ | ++ |
| pNS29HN | − | + | +++ |
| pNS29RHKRK | + | ++ | +++ |
Constructs indicated above are shown in Fig. 1 A and their construction is described in Materials and Methods. The relative fluorescence intensities distributed in different cell compartments, cytoplasm, nucleoplasm, and nucleoli, are presented from + to +++ ranking, and are based on 3–7 transfection experiments with each GFP fusion construct using the same set of parameters for measuring fluorescence intensity by confocal microscopy. Background autofluorescence of 3T3 fibroblasts is indicated by minus (−), whereas a slightly higher signal that may result from the expression of the fusion protein is indicated by minus and plus (−+).
The frequency of transfection, as measured by cells exhibiting fluorescence, was ∼2–10% with all constructs except for the pEGFP-Rpp38. This latter construct yielded transfectants at about one tenth the frequency of the other plasmids, presumably because RNase P incorporating this fusion protein is less active than in other cases. We note that for each construct tested, all transfected cells (i.e., cells exhibiting fluorescence) yielded the same phenotype as shown in Table .
Defining the Nucleolar Localization Domain of Rpp38
The carboxy terminal 24 amino acids of Rpp38 fused to GFP in pEGFP-Rpp38(260-283) (Fig. 1 A) are capable of introducing the reporter protein into the nucleoli of 3T3 fibroblasts (Table and data not shown). Nucleolar staining with GFP-Rpp38(260-283) was as intense as with GFP-Rpp38 and GFP-Rpp38(246-283) (compare relative intensities in Table ), an indication that the last 24–amino acid sequence of Rpp38, designated NS38, are sufficient for nucleolar localization of the reporter protein.
We investigated whether the lysine residues in NS38 were important for its function by amino acid substitution analysis. The intracellular distributions (cytoplasm, nucleoplasm, and nucleoli) as reflected by the fluorescence signals of several mutants shown in Fig. 1 A are also summarized in Table . Thus, pNS38KN, in which all the nine lysines in the NS38 sequence were substituted by similar, positively charged asparagine (N) residues, was introduced into 3T3 fibroblasts. The resultant fusion protein failed to enter the nucleoli and the fluorescent signal was detected in the cytoplasm as well as the nucleoplasm (Table ). When cells were transfected with pNS38KN23, in which lysines 2 and 3 in NS38 sequence were replaced by asparagines, or with pNS38KN45, in which lysines 4 and 5 were substituted, a marked decrease in the nucleolar fluorescence was measured (Table ). These latter two fusion proteins were also distributed evenly throughout the nucleus, when compared with the prominent concentration of GFP-Rpp38(260-283) in the nucleoli (Table ). Moreover, as in the case of GFP-Rpp38(260-283), NS38KN23 and NS38KN45 accumulated in the nucleoplasm (Table ), an indication that their nuclear retention was not completely abolished. Similar results were obtained with NS38KN59 in which lysines 5 and 9 were substituted (Table ). However, the nucleolar localization capability of NS38 was completely abolished when the double mutant NS38KN78, in which lysines at position 7 and 8 in NS38 were replaced by asparagines (Fig. 1 A), was introduced into cells (Table ). NS38KN78 was concentrated in the nucleoplasm rather than the cytoplasm when compared with NS38KN (Table ).
Next, we substituted alanine separately for each of the arginine (R13A), serine (S18A), threonine (T22A), or proline (P23A) residues in the NS38 sequence (Fig. 1 A) and tested the ability of these mutants to localize GFP to the nucleolus. The single mutants, R13A, S18A, and T22A, were found to have no profound effect on the nucleolar localization capability of NS38 (Table ). Therefore, phosphorylation of serine or threonine appears not to be an obligatory modification for NS38 function. The proline substitution to alanine (P23A), however, seemed rather to increase the ratio of the nucleolar to the nucleoplasmic staining when compared with the ratio obtained with the NS38 parental construct (Table ). The two prolines in the RKPP sequence of NS38, by contrast, had no critical role in nucleolar localization as corroborated by the ATΔPP construct in which the arginine and lysine (at position 5) were replaced with alanine and threonine, respectively, and the two consecutive proline residues were deleted from NS38 (Table ).
The findings described above show that lysine residues throughout the NS38 sequence are required for its nucleoplasmic retention and nucleolar localization, with the lysines at position 7 and 8 being most critical for its entry to the nucleolus.
GFP-Rpp38 Is Associated with Catalytically Active RNase P Complexes
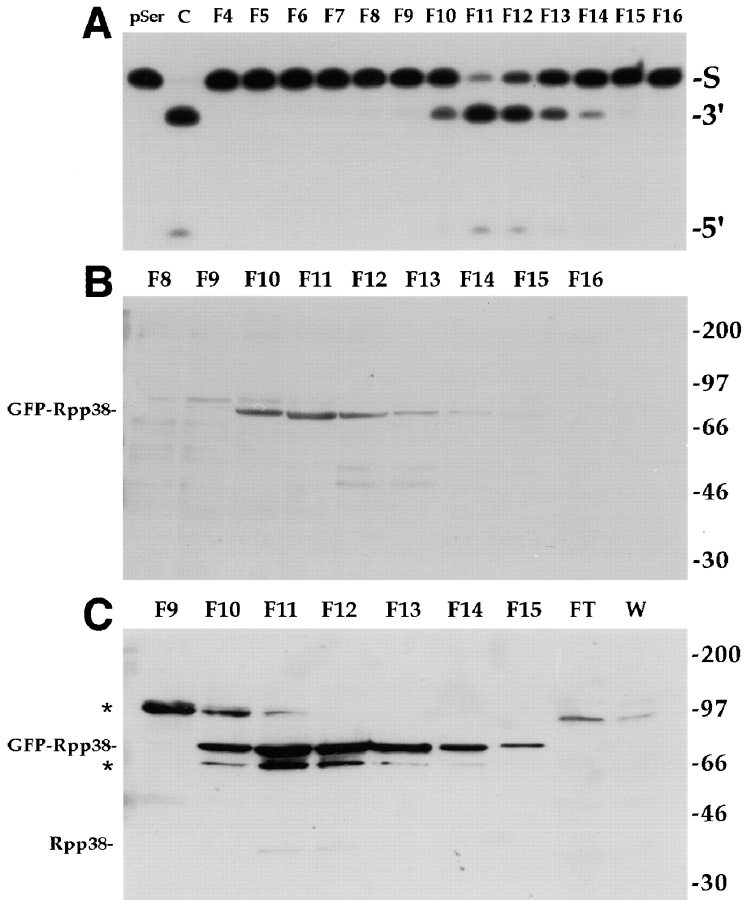
We obtained evidence that the GFP-Rpp38 fusion protein actually resides in a catalytically active RNase P complex. pEGFP-Rpp38, which expresses the neomycin resistance gene (G418 resistance), was used to establish stably transfected human embryonic kidney (HEK) 293 cells in culture. G418-resistant cell populations obtained in this manner exhibited fluorescent signals in the nucleoli and nucleoplasm, as judged by confocal microscopy (data not shown). To determine if GFP-Rpp38 expressed in these cells can be found in RNase P, S100 crude extracts from these stably transfected cells were fractionated on a DEAE-Sepharose anion exchange column. As determined by processing of a yeast tRNASer precursor, RNase P activity was eluted at 280–340 mM KCl (Fig. 5 A), a salt concentration shown previously to elute active RNase P from untransfected human cells from DEAE columns (Eder et al. 1997). When fractions across the peak of enzymatic activity were subjected to Western blot analysis using anti-GFP antibodies (see Materials and Methods), a protein of ∼75 kD that copurified with enzymatic activity was detected (Fig. 5 B). This protein corresponds to the GFP-Rpp38 fusion protein and apparently has an anomalous migration in SDS-PAGE, a property that is shared by several Rpp proteins including Rpp38 (Jarrous et al. 1998). When polyclonal anti-Rpp38 rabbit antibodies were used in Western blotting, the same 75-kD protein was detected (Fig. 5 C). A protein of ∼65 kD visible in the blot may be a truncated fragment of GFP-Rpp38. Neither flowthrough nor wash fractions from the column contained GFP-Rpp38 or endogenous Rpp38 protein (Fig. 5 C), an indication that both polypeptides were tightly bound to the column and eluted only with RNase P. At least as demonstrated in vitro, the expression of GFP-Rpp38 in human cells does not abolish RNase P function in tRNA processing, although constitutive expression of GFP-Rpp38 resulted in cell death after 10–15 passages in culture (data not shown).
Figure 5.
GFP-Rpp38 is associated with active RNase P. (A) S100 crude extracts obtained from of G418-resistant, stably transfected 293 HEK cell populations were fractionated on a DEAE-Sepharose anion exchange column (see Materials and Methods) and fractions eluted from the column were tested for RNase P activity using the yeast suppressor precursor tRNASer (S) as substrate (left lane). RNase P obtained from untransfected cells was used as a positive control (second lane from left). The cleavage products, 5′ leader sequence (5′) and mature tRNA (3′), were resolved in 8% polyacrylamide/7 M urea gel. Fraction numbers are indicated above the panel. Equal volumes from the fractions across the RNase P activity tested in A were separated in 12% SDS-PAGE and subjected to Western blot analysis using polyclonal anti-GFP rabbit antibodies (B) or affinity-purified, polyclonal anti-Rpp38 rabbit antibodies (C). FT and W in C stand for concentrated flowthrough and wash fractions of the DEAE column. Positions of GFP-Rpp38, endogenous Rpp38 protein, and protein size markers are shown. Cross-reaction with unknown proteins of M r ∼65 and ∼95 kD are indicated by asterisks.
Rpp29 Localizes a Reporter Protein to the Dense Fibrillar Component and Coiled Bodies
As with Rpp38, sequences of contiguous basic residues that may function in nuclear localization are found in Rpp29, another protein subunit of RNase P (Jarrous et al. 1999). 3T3 mouse fibroblasts were transfected with pEGFP-Rpp29, in which the open reading frame of the Rpp29 cDNA was fused in-frame with GFP (see Fig. 1 B). GFP-Rpp29 was localized in the nucleoplasm and exhibited very intense staining of nucleoli (Fig. 6, A–C). However, GFP-Rpp29 was not evenly distributed in the nucleoli but was concentrated in subregions inside these structures (Fig. 6, A–C). The smaller punctate stainings seen in the nucleoplasm may represent discrete structures other than nucleoli such as coiled bodies (see below). Indirect immunofluorescence analyses revealed that GFP-Rpp29 localized in nucleoli with B23 (Fig. 7, A–D). Moreover, endogenous Rpp29 colocalized with fibrillarin in untransfected fibroblasts (Fig. 7, E–G). The immunostains of Rpp29 and fibrillarin seen in nucleoli are strikingly similar (Fig. 7, E–G), suggesting that Rpp29 resides in the dense fibrillar component as fibrillarin.
Figure 6.
Subnucleolar localization of GFP-Rpp29 and identification of NS29. Swiss 3T3 fibroblasts were transiently transfected with pEGFP-Rpp29 (A–C) or pEGFP-Rpp29(52-85) (D–F). 48 h after transfection, localization of the fusion proteins was determined by confocal microscopy. DIC (left) GFP (middle), and overlay (right) images are shown. Arrows indicate nucleoli. Bars: C and F, 3.3 and 5 μm, respectively.
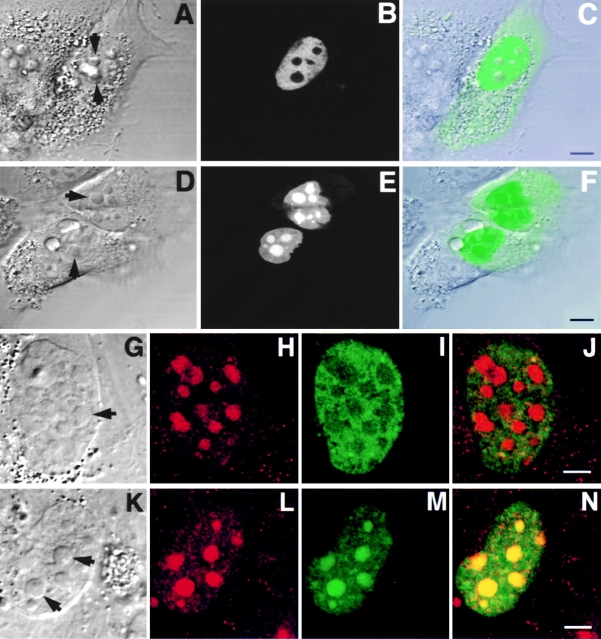
Figure 7.
Colocalization of Rpp29 with B23 and fibrillarin in the nucleolus. Swiss 3T3 fibroblasts were transfected with pEGFP-Rpp29. 48 h later, cells were fixed and subjected to immunostaining with an antibody against nucleolar protein B23 (A–D). Arrows indicate nucleoli. E and F show double immunofluorescence obtained with anti-Rpp29 and antifibrillarin antibodies, respectively, in untransfected 3T3 fibroblasts. G is an overlay of E and F, acquired at the same confocal plane using Texas red (red) or FITC (green) filters. An intense yellow color is seen in nucleoli. Bleedthrough between the two channels was completely eliminated. Bars: D and G, 2.5 and 5 μm, respectively.
As shown in Fig. 6D–F, the sequence responsible for the nucleolar localization of Rpp29 is located between positions 52 and 85 of this protein, as demonstrated by the use of pEGFP-Rpp29(52-85) (Fig. 1 B) in transfected fibroblasts. This domain was able to localize GFP to subnucleolar regions, but in a less distinct manner than the full-length Rpp29 protein. GFP-Rpp29(52-85) was almost exclusively retained in the nucleus and only background fluorescence was seen in the cytoplasm. Amino acids 63–85 of Rpp29 were still sufficient for nucleolar localization (Table ; construct pEGFP-Rpp29(63-85)). This sequence of 23–amino acid of Rpp29 is now designated NS29 (Fig. 1 B). Mutational analysis of NS29, summarized in Fig. 1 B and Table , showed that the KKKK residues in NS29 were required for efficient nucleolar localization but were not crucial for function, as determined through the use of NS29KN4 mutant. All the other multiple and single mutants tested, including the substitution of the RQRR residues to asparagines, had no dramatic effect on NS29 function (Table ). In fact, the RHKRK motif is sufficient for nucleolar entry (Table ). We concluded that NS29 and NS38 represent distinct domains required for nucleolar localization.
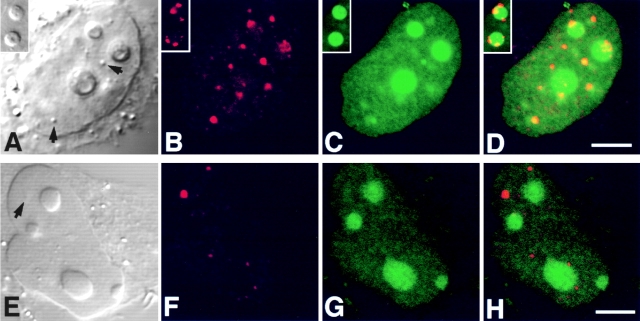
In HeLa cells, coiled bodies immunostained for p80-coilin contained GFP-Rpp29 (Fig. 8, A–D). In some transfected cells, however, coilin-immunostained structures that were on the periphery of nucleoli exhibited no intense signal of GFP-Rpp29 (Fig. 8, A–D, inserts). In contrast to GFP-Rpp29 and GFP-Rpp38, no prominent signal of GFP-Rpp14 fusion protein was seen in the coiled bodies of HeLa cells transiently transfected for 48 h with pEGFP-Rpp14 construct (Fig. 8, E–H). Whether Rpp14 requires a longer time to localize in coiled bodies, as was the case with its inefficient localization in the nucleolus (see below), remains unknown. A detailed kinetic study, however, is required to determine whether the several GFP-Rpp fusion proteins presented in this study enter the nucleoli first on their way to coiled bodies, as has been shown with Nopp140 (Isaac et al. 1998).
Figure 8.
GFP-Rpp29 is localized in nucleoli and coiled bodies. HeLa cells were transfected for 48 h with pEGFP-Rpp29 (A–D) or pEGFP-Rpp14 (E–H), and then immunostained for p80-coilin in indirect immunofluorescence analysis. DIC (A and E), p80-coilin (B and F, red), GFP (C and G, green), and the overlays of B over C and F over G are shown in D and H, respectively. Images in A–D and E–H were acquired at the same confocal plane. The GFP fluorescent signal in C and D was enhanced to highlight the punctate staining seen in the nucleoplasm. Coiled bodies are indicated by arrows. Inserts seen in A–D represent higher magnification of coilin-immunostained coiled bodies in the periphery to two nucleoli of HeLa cells expressing GFP-Rpp29. All images were obtained during the same experimental observation. Bars: D and H, 2.5 μm.
Nucleolar Localization of Rpp14, an RNase P Subunit, May Be Facilitated by a Piggyback Process
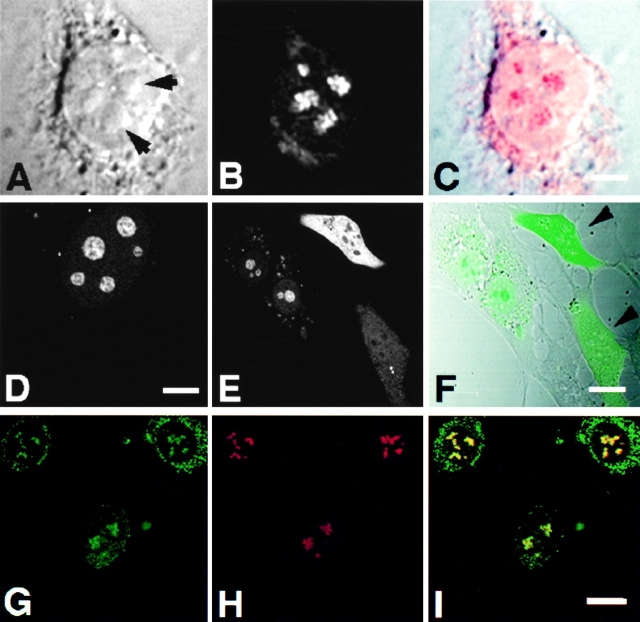
Indirect immunofluorescent analysis using affinity-purified, polyclonal antibodies against Rpp14 (Jarrous et al. 1999) showed localization of this RNase P subunit in the nucleolus of 3T3 fibroblasts (Fig. 9, A–C). Yet, Rpp14 has no sequences of basic residues typical of NLSs, in contrast to Rpp29 and Rpp38 (Jarrous et al. 1999). Nevertheless, Rpp14 fused to GFP was directed to subnucleolar regions in 3T3 fibroblasts transfected for 48 h with pEGFP-Rpp14 (Fig. 9, D–F). No prominent punctate staining was seen in the nucleoplasm, as was the case with Rpp29 and Rpp38. However, we found that GFP-Rpp14 was seen exclusively in the nucleolus only in cells producing low levels of this fusion protein, as reflected in the relatively weak fluorescent signals observed in transfected cells (Fig. 9E and Fig. F, note cell background). In transfected cells that showed more intense signals, comparable in their intensity to those reported here for Rpp38 and Rpp29, most of the GFP-Rpp14 synthesized was not transported to the nucleoli, but rather remained in the cytoplasm (Fig. 9E and Fig. F; two cells with stronger signals are indicated). Moreover, when cells were tested at earlier times (<24 h) after transfection, most of the GFP-Rpp14 stain was visible in the cytoplasm (data not shown). These observations suggest that, when compared with the rapid and efficient entry of Rpp38 and Rpp29 into the nucleolus, the localization process of Rpp14 under the same conditions is inefficient, and thus may require a limiting, endogenous factor. As demonstrated by co-localization analysis, GFP-Rpp14 was compartmentalized into subnucleolar regions occupied also by endogenous Rpp29 in transfected cells (Fig. 9, G–I), an indication that these two subunits colocalized in the dense fibrillar component.
Figure 9.
Subnucleolar localization patterns of Rpp14. 3T3 fibroblasts were subjected to indirect immunofluorescent analysis using anti-Rpp14 antibodies (A–C). C is an overlay of A and B. Fibroblasts transfected for 48 h with pEGFP-Rpp14 were examined under confocal microscope (D–F) before fixation and immunofluorescence analysis with anti-Rpp29 antibodies (G–I). F is an overlay of DIC (not shown) and E; two cells with high fluorescent signal are indicated by arrowheads. I is an overlay of G and H. Intense yellow color is seen in nucleoli. Bars: C and D, 3.3 μm; F and I, 10 μm.
Discussion
This study shows that the nucleolus of cultured mammalian cells serves as the major site of localization of several protein subunits of human RNase P. In contrast to the dispersed distribution of Rpp38 in nucleoli, the Rpp14 and Rpp29 subunits are localized in the dense fibrillar component. The differential pattern of nucleolar localization was also observed for these subunits when fused to GFP and expressed in living cells. These RNase P subunits, thus, define different sites that, in turn, may reflect distinct biological functions. This conclusion is supported further by our findings that at least two subunits, Rpp29 and Rpp38, are also found in functionally distinct organelles, the coiled bodies. The nucleolus and the coiled bodies appear to be involved in the biogenesis of the ribonucleoprotein RNase P and, therefore, in the process of maturation of tRNA precursors.
Molecular Aspects of Localization Domains in RNase P
Nucleolar localization of proteins usually involves multiple domains in targeting sequences that can interact with ribonucleic acids or with other proteins (Peculis and Gall 1992; Creancier et al. 1993; Yan and Melese 1993; Mears et al. 1995; Michael and Dreyfuss 1996; Antoine et al. 1997; Li 1997; Russo et al. 1997; Zirwes et al. 1997a,Zirwes et al. 1997b). Nucleolar localization domains of some proteins, such as nucleolin, p120 nucleolar protein, and ribosomal proteins L5 and L7a are not functional by themselves when transferred to a reporter protein; they require additional, noncontiguous domains for function (Schmidt-Zachmann and Nigg 1993; Valdez et al. 1994; Michael and Dreyfuss 1996; Russo et al. 1997). On the other hand, NS38 and NS29 are functional and transferable. However, these two domains seem not to be sufficient for targeting a reporter protein to the coiled bodies as well.
NS38 has no arginine- or arginine/glycine–rich motifs (Burd and Dreyfuss 1994), as has been found in domains in nucleolin (C23) and in the human immunodeficiency virus Tat protein that may facilitate RNA-binding and/or protein–protein interactions (Dang and Lee 1989; Schmidt-Zachmann and Nigg 1993; Mears et al. 1995; Bouvet et al. 1998). The single arginine residue found in NS38 has no essential role either in the nucleoplasmic retention or in the nucleolar localization capability of this domain. Lysine residues at different positions throughout the NS38 sequence, instead, are required for efficient nucleolar localization. Adjacent lysines at positions 7 and 8, but not at positions 2 and 3, have a critical role in NS38 function. Numerous KKX repeats are found in several protein subunits of yeast nuclear RNase P (Chamberlain et al. 1998), but as in many other cases of nucleolar proteins such repeats were proved nonessential for nucleolar targeting (Gautier et al. 1997). NS38 shows no identity at the primary amino acid sequence to NS29. It is thus likely that structural features and the placement in space of side chains of both hydrophobic and charged amino acids (lysines) determine the function of these sequences.
Both NS29 and NS38 act early and efficiently to introduce a reporter protein to the nucleoli of mouse and human cultured cells. Similar conclusions were made for the full-length proteins, Rpp29 and Rpp38. In contrast, Rpp14 entry to the nucleolus seems slow and limited. Rpp14, which lacks any basic residues typical of nuclear or nucleolar targeting domains, may require other proteins that occur in limited amounts in the cell for its nucleolar transport.
Furthermore, we were able to show that the nucleolar localization processes of Rpp subunits are dependent on ongoing transcription in functional, intact nucleoli. Thus, selective inhibition of rRNA transcription by a low concentration of actinomycin D (0.2 μg/ml; Pombo et al. 1999) leads to disintegration of the nucleoli and to dispersed nucleoplasmic staining by Rpp29, Rpp38, or their nucleolar localization domains fused to GFP (data not shown). Inhibition of protein synthesis by cycloheximide, by contrast, seems to have no effect on the nucleolar localization properties of these subunits.
Localization Sites of RNase P and Biological Functions
As judged by RNA hybridization analysis in situ, most of the RNA subunit of the yeast nuclear RNase P is localized in the nucleolus with some unprocessed tRNA precursors that contain 5′ leader sequences (Bertrand et al. 1998). In contrast, the majority of the human RNase P RNA is concentrated in the nucleoplasm rather than the nucleolus (Matera et al. 1995; Lee et al. 1996; Jacobson et al. 1997; Wolin and Matera 1999). Moreover, H1 RNA that was microinjected to the nucleoplasm only transiently enters the dense fibrillar component of the nucleolus before it is redistributed in the nucleoplasm (Jacobson et al. 1997). Our study now shows, using both indirect immunofluorescence and cell transfection analyses, that several protein subunits of human RNase P reside in the nucleolus. Since the estimated copy numbers of RNase P RNA and RNase MRP RNA in a metazoan cell are ∼2 × 105 and ∼105, respectively (Yu et al. 1999), there are at least two explanations of these differences in the location of the RNA and the protein subunits: newly synthesized H1 RNA enters the nucleolus for assembly with the Rpp subunits before it exits to the nucleoplasm, or that the nucleolus acts as a sequestration compartment (Bachant and Elledge 1999) for several Rpp subunits that can be recruited to other nucleoplasmic sites, where H1 RNA exists, to form an active RNase P complex under certain physiological conditions. The localization of these subunits in the coiled bodies supports the idea that these organelles may be involved in sorting and transport of RNase P and RNase MRP components from the nucleolus to other destinations and vice versa.
Finally, RNA and protein subunits of RNase P and RNase MRP colocalize in the dense fibrillar component of the nucleoli and both utilize a common conserved RNA structural element, the P3 domain (Forster and Altman 1990), for their nucleolar entry (Jacobson et al. 1995, Jacobson et al. 1997). The localization pattern of Rpp29 in the nucleolus itself suggests that this subunit localizes RNase P and RNase MRP to the dense fibrillar component in which transcription and early rRNA processing events take place (Shaw et al. 1995). Colocalization studies with Nopp140 and B23, the latter is found in the granular component in addition to the dense fibrillar component (Biggiogera et al. 1990), indicate that Rpp38 may also reside in other compartments known as sites of preribosome assembly (Shaw et al. 1995). Whether these compartments are involved in the processing of some precursor tRNAs remains unknown. However, a common molecular process may govern the localization and assembly of RNase P and RNase MRP to ensure the coordinated processing of stable RNA in mammalian cells.
Acknowledgments
We thank Joel Rosenbaum and Mark Mooseker (both from Yale University) for reading the manuscript and for valuable suggestions and Paul S. Eder (University of Pennsylvania) for advice in immunofluorescence. We thank Joseph Gall (Carnegie Institution, Baltimore), Thomas Meier (Albert Einstein College of Medicine), Harris Busch and Yong Ren (both from Baylor College of Medicine) for antibodies against p80 coilin, Nopp140, and fibrillarin, respectively, and our colleagues, especially Cecilia Guerrier-Takada and Petur H. Peterson for technical assistance.
N. Jarrous was supported by a postdoctoral fellowship from Innovir Laboratories. J.S. Wolenski is supported by a National Science Foundation grant BIR-9601664. This work was supported by the Human Frontiers Science Program grant RG O2N1 1997M and the U.S. Public Health Service grant GM-19422 to S. Altman.
Footnotes
1.used in this paper: DIC, differential interference contrast; GFP, green fluorescent protein; HEK, human embryonic kidney; NLS, nuclear localization sequence; RNase MRP, ribonuclease mitochondrial RNA processing; RNase P, ribonuclease P; Rpp, RNase P protein
References
- Andrade L.E.C., Tan E.M., Chan E.K.L. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc. Natl. Acad. Sci. USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine M., Reimers K., Dickson C., Kiefer P. Fibroblast growth factor 3, a protein with dual subcellular localization, is targeted to the nucleus and nucleolus by the concerted action of two nuclear localization signals and a nucleolar retention signal. J. Biol. Chem. 1997;272:29475–29481. doi: 10.1074/jbc.272.47.29475. [DOI] [PubMed] [Google Scholar]
- Bachant J.B., Elledge S.J. Mitotic treasures in the nucleolus. Nature. 1999;398:757–758. doi: 10.1038/19641. [DOI] [PubMed] [Google Scholar]
- Bauer D.W., Murphy C., Wu Z., Wu C.H., Gall J.G. In vitro assembly of coiled bodies in Xenopus egg extract. Mol. Biol. Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E., Houser-Scott F., Kendall A., Singer R.H., Engelke D.R. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggiogera M., Burki K., Kaufmann S.H., Shaper J.H., Gas N., Amalric F., Fakan S. Nucleolar distribution of proteins B23 and nucleolin in mouse preimplantation embryos as visualized by immunoelectron microscopy. Development. 1990;110:1263–1270. doi: 10.1242/dev.110.4.1263. [DOI] [PubMed] [Google Scholar]
- Bouvet P., Diaz J.J., Kindbeiter K., Madjar J.J., Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Chamberlain J.R., Lee Y., Lane W.S., Engelke D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creancier L., Prats H., Zanibellato C., Amalric F., Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol. Biol. Cell. 1993;4:1239–1250. doi: 10.1091/mbc.4.12.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V., Lee W.M. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J. Biol. Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- Eder P.S., Kekuda R., Stolc V., Altman S. Characterization of two scleroderma autoimmune antigens that copurify with the ribonucleoprotein ribonuclease P. Proc. Natl. Acad. Sci. USA. 1997;91:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A.C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Tsvetkov A., Wu Z., Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev. Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gautier T., Berges T., Tollervey D., Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Link between aging and the nucleolus. Genes Dev. 1997;11:2449–2455. doi: 10.1101/gad.11.19.2449. [DOI] [PubMed] [Google Scholar]
- Isaac C., Yang Y., Meier U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M.R., Cao L.G., Wang Y.L., Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M.R., Cao L.G., Taneja K., Singer R.H., Wang Y.L., Pederson T. Nuclear domains of the RNA subunit of RNase P. J. Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jarrous N., Eder P.S., Guerrier-Takada C., Hoog C., Altman S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- Jarrous N., Eder P.S., Wesolowski D., Altman S. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA. 1999;5:153–157. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lee B., Matera A.G., Ward D., Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolusa possible coordinate role in ribosome biogenesis. Proc. Natl. Acad. Sci. USA. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.P. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J. Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Pluk H., van Venrooij W.J., Seraphin B. hPop1an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Frey M.R., Margelot K., Wolin S.L. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract–binding protein, hnRNP I. J. Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears W.E., Lam V., Rice S.A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U.T., Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Melese T., Xue Z. The nucleolusan organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- Melton D.A., Cortese R. Transcription of cloned tRNA genes and the nuclear partitioning of a tRNA precursor. Cell. 1979;18:1165–1172. doi: 10.1016/0092-8674(79)90229-0. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Dreyfuss G. Distinct domains in ribosomal protein L5 mediate 5 S rRNA binding and nucleolar localization. J. Biol. Chem. 1996;271:11571–11574. doi: 10.1074/jbc.271.19.11571. [DOI] [PubMed] [Google Scholar]
- Peculis B.A., Gall J.G. Localization of the nucleolar protein NO38 in amphibian oocytes. J. Cell Biol. 1992;116:1–14. doi: 10.1083/jcb.116.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3877. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H., van Eenennaam H., Rutjes S.A., Pruijn G.J., van Venrooij W.J. RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A., Jackson D.A., Hollinshead M., Wang Z., Roeder R.G., Cook P.R. Regional specialization in human nucleivisualization of discrete sites of transcription by RNA polymerase III. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo G., Ricciardelli G., Pietropaolo C. Different domains cooperate to target the human ribosomal L7a protein to the nucleus and to the nucleoli. J. Biol. Chem. 1997;272:5229–5235. doi: 10.1074/jbc.272.8.5229. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M.S., Nigg E.A. Protein localization to the nucleolusa search for targeting domains in nucleolin. J. Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- Shaw P.J., Jordan E.G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Shaw P.J., Highett M.I., Beven A.F., Jordan E.G. The nucleolar architecture of polymerase I transcription and processing. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:2896–2906. doi: 10.1002/j.1460-2075.1995.tb07289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez B.C., Perlaky L., Henning D., Saijo Y., Chan P.K., Busch H. Identification of the nuclear and nucleolar localization signals of the protein p120. Interaction with translocation protein B23. J. Biol. Chem. 1994;269:23776–23783. [PubMed] [Google Scholar]
- Wolin S.L., Matera A.G. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- Yan C., Melese T. Multiple regions of NSR1 are sufficient for accumulation of a fusion protein within the nucleolus. J. Cell Biol. 1993;123:1081–1091. doi: 10.1083/jcb.123.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.-T., E.C. Scharl, C.M. Smith, and J.A. Steitz. 1999. The growing world of small nuclear ribonucleoproteins. In The RNA World. Second edition. Cold Spring Harbor Press, Cold Spring Harbor, New York. 487–524.
- Zirwes R.F., Kouzmenko A.P., Peters J.M., Franke W.W., Schmidt-Zachmann M.S. Topogenesis of a nucleolar proteindetermination of molecular segments directing nucleolar association Mol. Biol. Cell. 8 1997. 231 248a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirwes R.F., Schmidt-Zachmann M.S., Franke W.W. Identification of a small, very acidic constitutive nucleolar protein (NO29) as a member of the nucleoplasmin family Proc. Natl. Acad. Sci. USA. 94 1997. 11387 11392b [DOI] [PMC free article] [PubMed] [Google Scholar]