Abstract
New evidence supports the idea of a nonmicrotubule spindle matrix, but the debate about the reality of this structure continues.
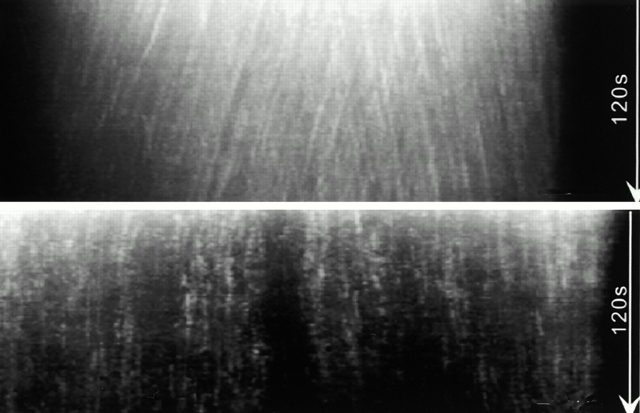
Motors and movement go hand in hand, and microtubule-based motors in the spindle are no exception. Thus, the findings of Kapoor and Mitchison on page 1125 of this issue come as a surprise. Put simply, they claim that the Eg5 kinesin motor stays put in spindles assembled in frog egg extracts (Fig. 1), despite the constant movement of microtubules in a polewards flow called flux.
Figure 1.
New evidence for a possible spindle matrix. Speckles of polymerized tubulin (top) move over time (top to bottom), whereas speckles of the kinesin motor Eg5 (bottom) stay stationary relative to the spindle. Thus, the Eg5 may be tethered by a spindle matrix.
One possible explanation for this behavior is that Eg5 is walking along microtubules with a motion that is equal in rate but opposite in direction to flux. In this way, motors could appear stationary in a relative sense. But when Kapoor and Mitchison add monastrol, an Eg5 inhibitor, the Eg5 distribution remains unchanged even as flux continues.
“Our observation is that Eg5 is static in the spindle, whereas tubulin is moving,” says Kapoor. “The question is how do we interpret this.”
Their provocative answer, for now, is that Eg5 is tethered by a spindle matrix that gives spindle motors something to push against (see Scholey et al., 2001, for a review of evidence regarding a possible spindle matrix). Not everyone agrees. “The spindle matrix has been an unproven entity so there are going to be believers and nonbelievers,” says Duane Compton (Dartmouth Medical School, Hanover, NH). “I'm on the fence.”
What's that stuff?
Of the numerous candidates for the title of spindle matrix, many cover only a particular region of the spindle. Hints at the existence of one of those regional matrices arose decades before the identification of tubulin, arguably with the observations of Karl Belar in the 1920s (Belar, 1929). He saw something dark and mysterious in the center of the spindle, named it the “pushing body” (Stemmkörper), and stated that it was responsible for pushing spindle poles apart during the spindle-lengthening process of anaphase B. The physical manifestation of this proposed pushing activity was apparent as electron-dense material in later electron micrographs (Paweletz, 1967) in a structure that was probably what we now call the midbody (Margolis and Andreassen, 1993).
“There is a whole lot of stuff between the microtubules in the middle region of the spindle,” says Richard McIntosh (University of Colorado, Boulder, CO). “I would assume it is the very same stuff that the Mitchison group is now looking at.”
The midbody covers the central region of the spindle where microtubules from opposite poles overlap and interdigitate. As the extent of that overlap decreases during anaphase B, the midbody gets thinner. It has been suggested that the midbody acts to push the poles apart, stabilize the central spindle, induce the cleavage furrow, or serve as a dumping ground for mitotic proteins that are no longer needed.
In the same region of the metaphase spindle is the corona, a set of filaments extending out from kinetochores that may help kinetochores to capturemicrotubules and generate force. In the green alga Oedogonium, these filaments can extend several micrometers into the nucleoplasm (Schibler and Pickett-Heaps, 1980; Pickett-Heaps and Carpenter, 1993), but in most other organisms they are only a few hundred nanometers long.
At the two extremes of the spindle is perhaps the best candidate for the title of a spindle matrix protein: NuMA. Dynein motors transport NuMA protein to the spindle poles where it forms an extensive structure that is essential for spindle formation in frog egg extracts (Merdes et al., 1996; Dionne et al., 1999). NuMA may use dynein only for transport to the spindle pole before forming a matrix via self-association. Alternatively, dynein may drag polewards both NuMA and the microtubules that are bound to NuMA, thus focussing the spindle pole.
The NuMA matrix can be sizeable (extending up to two or three micrometers from the pole in a spindle that is perhaps ten micrometers long), but like the midbody and corona it does not cover the whole spindle. “A NuMA-like matrix is probably going to be linked to the poles,” says Compton. “It is probably not analogous to what an Eg5 attachment matrix might be.”
A matrix to cover it all
Kapoor and Mitchison observed stationary Eg5 over the entire spindle, so any matrix explanation of their results would have to invoke a matrix that spans from pole to pole. This kind of matrix “is a very interesting idea,” says Compton, “and I think there are certain properties of spindles, notably flux, that would require a matrix.”
Microtubule flux involves polewards microtubule movement without shortening: tubulin subunits are added to the microtubule ends in the center of the spindle and depolymerized from the microtubule ends near the poles. The function of flux is not known, since the known treatments that inhibit flux are sufficiently nonspecific that they also destroy spindle integrity.
The problem with flux comes not with the providers of force—there are plenty of motors around—but with what those motors are using to brace themselves against. “I have a hard time understanding the balance of forces when the tracks on which you are pushing are constantly slipping,” says Kapoor. A matrix is one possible explanation for such a structural support. A local matrix such as NuMA is a start, especially for flux, but for chromosome movement there must be a mechanism ensuring that chromosomes move to the poles rather than poles moving to the chromosomes.
Jonathan Scholey (University of California, Davis, CA) thinks this problem can be solved without the need for a special matrix. “People have been pushing the idea of a spindle matrix for decades,” he says. “It might exist, but there is no hard evidence.” For the structural support, he says, “an extensively cross-linked lattice of microtubules could function like a spindle matrix, but there is no need to refer to it as a spindle matrix.”
Scholey is influenced by his findings with motors such as the fly protein KLP61F, which like Eg5 is a bipolar kinesin in the BimC class. KLP61F acts as a microtubule cross-linker that may bridge microtubules throughout the spindle and push apart antiparallel microtubules within interpolar microtubule bundles crossing the spindle midzone (Sharp et al., 1999). If this pushing force can maintain a set distance between poles, the kinetochore microtubules could work against the framework of cross-linked interpolar microtubule bundles while reeling in the chromosomes during anaphase. These bundles may have to work as part of an ever-changing structure, as Kapoor notes that no one has observed a stable, static subset of microtubules.
All this talk about forces is not new: the debate about a spindle matrix has always been linked to arguments about how force is generated in the spindle. Juan Subirana, in one of the first papers suggesting an all-encompassing spindle matrix, recognized that microtubules could act as struts (Subirana, 1968). But, at the time, he and others believed that kinetochores were largely passive, so he proposed that matrix-embedded motors would be needed to drive chromosome movement to the poles.
Meanwhile, Pickett-Heaps et al. (1982) proposed that chromosome movement to the metaphase plate stretched an elastic matrix that, at the onset of anaphase, recoiled to drag in the chromosomes—the reverse of the original Stemmkörper idea. Mitchison is interested in how a similar concept may apply to tubules derived from the endoplasmic reticulum. “They are mostly on the outside of spindles,” he says, “but they stretch from pole to pole, and so might act as a kind of spring compressing the spindle.”
Beat on the cell
Current theory may not have a need for a matrix covering the whole spindle, but that doesn't mean that it doesn't exist. Pickett-Heaps et al. (1982) summarize a variety of experiments in which fibrous remnants were observed in sea urchin spindles after microtubule extraction. Unfortunately, says McIntosh, in the fixation leading up to the electron micrography used in these experiments “the treatments were barbarous. You only see [the fibers] when you beat on the cell in various ways.” Based on this evidence, he says, “I would put no great confidence in the idea of a spindle matrix.”
Rapid freezing techniques (rather than chemical fixation) have reduced such EM artifacts but can result in the disappearance of some known matrices. The solution, say nearly all involved, is the isolation of specific matrix proteins, if they exist.
The direct approach to isolation has proven less than useful. “The spindle is a sponge,” says McIntosh. So many proteins glom onto it that, in previous attempts, “the number of components [copurifying with spindles] was limited only by the resolving power of the analytical method.”
Luck has come to the rescue. Candidate spindle matrix proteins have been identified thanks to antibodies originally raised against a nuclear transport targeting sequence (Paddy and Chelsky, 1991), sea urchin axonemes (Steffen and Linck, 1992), or a portion of the neurogenic protein Notch (Walker et al., 2000, but note that the final serum recognizes an antigen unrelated to Notch). Unfortunately, two of the isolated proteins (Spoke [Paddy and Chelsky, 1991] and Tektin Tektin [Steffen and Linck, 1992]) have disappeared from the mitosis literature.
What's in a name?
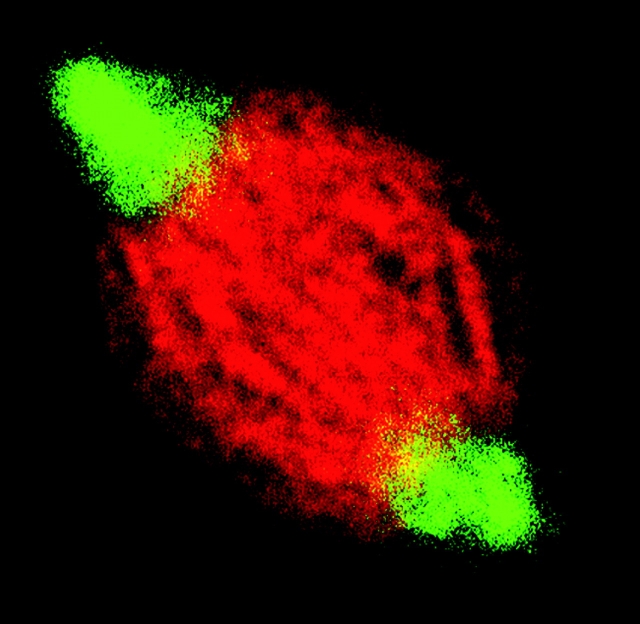
In one of the more audacious acts of nomenclature last year, Walker et al. (2000) dubbed their matrix candidate Skeletor. Even before a microtubule-based spindle forms, and after microtubules are depolymerized with drugs, Skeletor staining shows a provocative spindle-like shape (Fig. 2).
Figure 2.
Skeletor (red) forms a structure in the nucleus even before the formation of an extensive microtubule-based spindle (green). See http://www.jcb.org/cgi/content/full/151/7/1401/DC1 for related videos.
The Skeletor experiments were performed in fly embryos that retain nuclear envelopes well into mitosis. Skeletor staining fills a shape similar to that outlined by the nuclear envelope, but Kristen Johansen (Iowa State University, Ames, IA) says that Skeletor is not associated merely with nuclei or nuclear envelopes. She notes that fibrous Skeletor staining is visible throughout the spindle (not just next to the envelope) and is oriented from pole to pole even before there is a significant number of microtubules in the nucleus. “If this were simply the envelope constraining the protein, you wouldn't see this kind of positioning information,” she says.
What is lacking in last year's paper is functional evidence that Skeletor does anything. Johansen is trying hard to get such evidence using either mutants or double-stranded RNA, and she has isolated two proteins that interact with Skeletor, one of which, she says, looks structural. “Once a more structural protein is identified along with a mutant phenotype, that should satisfy the skeptics,” she says.
Motor evidence
Meanwhile, Johansen is excited about the latest Eg5 data because it introduces a motor protein into the debate. That has happened before, notably in a paper from Scholey and colleagues, in which they observed kinesin immobilized in spindle regions even after extraction of microtubules (Leslie et al., 1987). But in subsequent work, Scholey found that the kinesin was anchored to membranous structures that probably aid in repair of plasma membrane damage (Wright et al., 1991). With regard to a possible spindle matrix, says Scholey, “the whole body of work is a red herring.”
But this time around, the motor, Eg5, has a known role in maintaining the bipolarity of the spindle. So how, according to the matrix skeptics, does Eg5 remain stationary? In the middle of the spindle, Eg5, which is known to multimerize, could grab onto antiparallel microtubules that are fluxing in opposite directions. But it is harder to understand how Eg5 could stay in place outside of this central region of microtubule overlap without invoking a spindle matrix.
There are caveats of course—notably that the chemically labeled Eg5 could be an inactive motor with the microtubule gliding that Kapoor and Mitchison observe in vitro being provided by a subset of unlabeled protein. Kapoor believes this is not the case based on localization evidence. The labeled protein localizes correctly to the spindle, unlike other recombinant versions of Eg5 tested previously, and the localization is perturbed by the same treatments that perturb endogenous Eg5. “To me that is the best evidence that this isn't chunks of protein getting stuck,” says Kapoor.
Kapoor's experiments do not speak to the shape or structure of any potential matrix. He thinks the matrix could be something as simple as oligomerized Eg5, perhaps assembling into short filaments that crosslink microtubules, in an exaggerated version of the Scholey model. Or Eg5 might interact with something like Skeletor.
The nature of the filaments, if they exist, may come to light with immuno EM. Will they look like the spindle remnants seen in earlier micrographs of sea urchins, which for now are largely dismissed as artifacts? If so, it would be ironic indeed. The existence of the microtubule spindle was inferred in 1876, but its fibers were considered a fixation artifact until the 1920s (Paweletz, 2001). Perhaps the nonmicrotubule spindle matrix will make a similar comeback.
Acknowledgments
Thanks to Tarun Kapoor for supplying references, and to J. Richard McIntosh for help with the historical literature.
References
- Belar, K. 1929. Beiträge zur Kausalanalyse der Mitose. Wilhelm Roux Archiv. Entwicklungsmech. Org. 118:359–484. [DOI] [PubMed] [Google Scholar]
- Dionne, M.A., L. Howard, and D.A. Compton. 1999. NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil. Cytoskeleton. 42:189–203. [DOI] [PubMed] [Google Scholar]
- Kapoor, T.M., and T.J. Mitchison. 2001. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154:1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, R.J., R.B. Hird, L. Wilson, J.R. McIntosh, and J.M. Scholey. 1987. Kinesin is associated with a nonmicrotubule component of sea urchin mitotic spindles. Proc. Natl. Acad. Sci. USA. 84:2771–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis, R.L., and P.R. Andreassen. 1993. The telophase disc: its possible role in mammalian cell cleavage. Bioessays. 15:201–217. [DOI] [PubMed] [Google Scholar]
- Merdes, A., K. Ramyar, J.D. Vechio, and D.W. Cleveland. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. [DOI] [PubMed] [Google Scholar]
- Paddy, M.R., and D. Chelsky. 1991. Spoke: a 120-kD protein associated with a novel filamentous structure on or near kinetochore microtubules in the mitotic spindle. J. Cell Biol. 113:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweletz, N. 1967. Zur Funktion des “Fleming-Korpers” bei der Teilung tierscher Zellen. [On the function of the “Flemming body” during division of animal cells.] Naturwissenschaften. 54:533–535. [DOI] [PubMed] [Google Scholar]
- Paweletz, N. 2001. Walther Flemming: pioneer of mitosis research. Nat. Rev. Mol. Cell. Biol. 2:72–75. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D., D.H. Tippit, and K.R. Porter. 1982. Rethinking mitosis. Cell. 29:729–744. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D., and J. Carpenter. 1993. An extended corona attached to metaphase kinetochores of the green alga Oedogonium. Eur. J. Cell Biol. 60:300–307. [PubMed] [Google Scholar]
- Schibler, M.J., and J.D. Pickett-Heaps. 1980. Mitosis in Oedogonium: spindle microfilaments and the origin of the kinetochore fiber. Eur. J. Cell Biol. 22:687–698. [PubMed] [Google Scholar]
- Scholey, J.M., G.C. Rogers, and D.J. Sharp. 2001. Mitosis, microtubules, and the matrix. J. Cell Biol. 154:261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., K.L. McDonald, H.M. Brown, H.J. Matthies, C. Walczak, R.D. Vale, T.J. Mitchison, and J.M. Scholey. 1999. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144:125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, W., and R.W. Linck. 1992. Evidence for a non-tubulin spindle matrix and for spindle components immunologically related to tektin filaments. J. Cell Sci. 101:809–822. [DOI] [PubMed] [Google Scholar]
- Subirana, J.A. 1968. Role of spindle microtubules in mitosis. J. Theor. Biol. 20:117–123. [DOI] [PubMed] [Google Scholar]
- Walker, D.L., D. Wang, Y. Jin, U. Rath, Y. Wang, J. Johansen, and K.M. Johansen. 2000. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J. Cell Biol. 151:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, B.D., J.H. Henson, K.P. Wedaman, P.J. Willy, J.N. Morand, and J.M. Scholey. 1991. Subcellular localization and sequence of sea urchin kinesin heavy chain: evidence for its association with membranes in the mitotic apparatus and interphase cytoplasm. J. Cell Biol. 113:817–833. [DOI] [PMC free article] [PubMed] [Google Scholar]