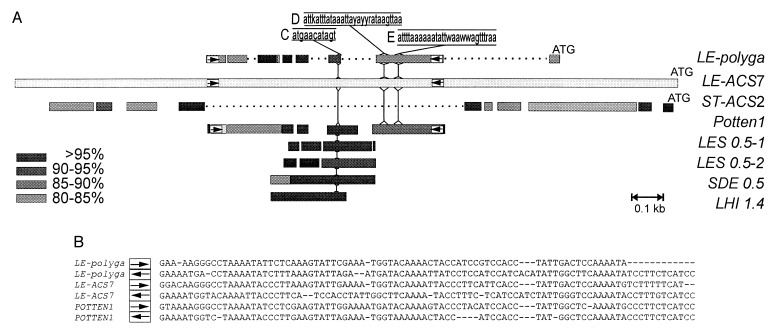
Figure 6.
Comparison of the LE-ACS7 promoter with other plant promoters and with Sol3 elements. (A) The bars represent alignments of DNA sequences as revealed by a gapped blast search (44). The sequence names are printed on the right of each bar, and the distance scale (0.1 kb) is shown. The promoter of LE-ACS7 is depicted as a light grey bar, beginning on the left with nt 1 and ending on the right with the initiation codon ATG, i.e., nt 2,466 of Fig. 1. Blocks of DNA sequence identity to the LE-ACS7 promoter are aligned above and below the LE-ACS7 promoter, at various grey shades to represent their percentage of identity to the corresponding regions of the LE-ACS7 promoter, as indicated in the legend. Blocks with identities of <80% to the LE-ACS7 promoter were not plotted. Gaps (… .) were inserted in the promoters of ST-ACS2 and LE-polyga. White boxes with arrows indicate the positions of the long terminal inverted repeats characteristic of Sol3 transposons. The position of the nuclear protein binding DNA sequences C, D, and E in the positive regulatory region of the PG promoter (nucleotides −412 to −806) (31) and their consensus sequence with LE-ACS7 are shown. Vertical lines show where these regions are located on the LE-ACS7 promoter and on various Sol3 elements. The Sol3 DNA sequences used are: Solanum tuberosum (Potten1) GenBank accession no. U91987; Solanum demissum (SDE 0.5) accession no. U91992; Lycopersicon esculentum (LES 0.5–1), accession no. U91989; (LES 0.5–2), accession no. U91990; Lycopersicum hirsutum (Lhi 1.4), accession no. U91988. (B) Alignment of the long terminal inverted repeats of LE-polyga, LE-ACS7, and Potten1.