The expression, purification, and crystallization in space group P212121 of the complex HasA-HasR from S. marcescens are reported. Diffraction data have been collected and processed to 6.8 Å.
Keywords: HasR, HasA, TonB, haem acquisition, outer-membrane receptor
Abstract
Serratia marcescens is able to acquire iron using its haem-acquisition system (‘has’), which contains an outer membrane receptor HasR and a soluble haemophore HasA. After secretion, HasA binds free haem in the extracellular medium or extracts it from haemoproteins and delivers it to the receptor. Here, the crystallization of a HasA–HasR complex is reported. HasA and HasR have been overexpressed in Escherichia coli and the complex formed and crystallized. Small platelets and bunches of needles of dimensions 0.01 × 0.1 × 1 mm were obtained. A native data set has been collected to 6.8 Å.
1. Introduction
Iron is required in many proteins as a cofactor or for local structural stabilization. Despite being abundant in the natural environment, iron is only available to bacteria with difficulty owing to its predominant existence as insoluble FeIII under oxidative conditions. In order to survive in eukaryotes, pathogenic bacteria need to acquire bound iron from transferrin, lactoferrin (Hentze et al., 2004 ▶), haemoglobin, myoglobin or haemopexin.
Gram-negative bacteria have developed systems for the uptake and transport of iron through the outer membrane. When living in the environment, bacteria secrete small water-soluble molecules which chelate FeIII with high affinity (Neilands, 1995 ▶). Specific outer membrane receptors bind the iron-loaded siderophores and translocate them through the outer membrane using chemiosmotic energy.
Under iron-starvation conditions, Serratia marcescens cells secrete a haem-binding protein, HasA (Swiss-Prot Q54450), which is able to extract haem from haemoglobin (K a = 5 × 1010 M −1; Létoffé et al., 1994 ▶; Wandersman & Stojiljkovic, 2000 ▶; Deniau et al., 2003 ▶). Subsequently, HasA is bound by a specific outer membrane receptor HasR (Swiss-Prot Q79AD2) and translocation of haem through the outer membrane takes place driven by chemiosmotic energy. The latter is transduced to HasR in the outer membrane by the mechanism described for siderophore receptors by a cytoplasmic membrane complex TonB–ExbB–ExbD, although S. marcescens has its own TonB-like protein in HasB (Létoffé et al., 1999 ▶, 2004 ▶; Postle & Kadner, 2003 ▶).
The crystal structure of holoHasA has been solved (PDB code 1b2v; Arnoux et al., 1999 ▶) and shows the haem iron coordinated by His32 and Tyr75 in two surface-exposed loops. HasR forms complexes with both haem-free apoHasA and holoHasA (Ghigo et al., 1997 ▶). The interaction is also of high affinity (K a = 5 × 109 M −1), is mediated by two β-strands of HasA and occurs independently of TonB (Létoffé et al., 2001 ▶, 2003 ▶). Bound holoHasA delivers its haem to HasR, while the apoHasA is recycled.
The structure of HasR is unknown. Sequence alignment of several homologues indicates a β-barrel protein with an N-terminal plug domain similar to other siderophore receptors of known structure (Ferguson et al., 2002 ▶). The barrel is inserted in the outer membrane, with loops liganding the β-strands in the extracellular medium. The plug is inserted into the barrel. In the case of FhuA, an active receptor can be made from separately synthesized plug and barrel domains (Braun et al., 2003 ▶). HasR has an N-terminal extension of about 80 residues that is absent in most siderophore receptors except for FecA and FpvA and is involved in transcriptional signalling (Enz et al., 2003a ▶,b ▶). Binding of holoHasA to the outer membrane receptor HasR inactivates the antisigma factor, turning on the sigma factor which promotes has transcription. Deletion of the N-terminal extension abolishes the inducing activity but not the transport activity (Biville et al., 2004 ▶). Two histidines [one in the plug, His189 (numbered from the first residue of the mature protein), and one in the barrel, His603] are conserved among HasR orthologues. In a similar receptor, HemR from Yersinia enterocolitica, mutations showed that they are involved in haem transport (Bracken et al., 1999 ▶).
In the absence of HasA, haem acquisition is supported at concentrations of haem in the surrounding medium above 10−6 M. When HasA is present, haem acquisition still takes place at concentrations as low as 10−8 M, but requires higher energy driven by higher TonB-complex concentration (Létoffé et al., 2004 ▶). HasA thus plays the role of a haem scavenger around the receptor. These two pathways raise structural and functional questions about the receptor. Currently, no structure of an outer membrane haem receptor is known, so that the structures of various siderophores and vitamin B12 receptors (Ferguson et al., 1998 ▶, 2002 ▶; Chimento et al., 2003 ▶) have to be used as models for them in spite of large differences in function.
In this paper, we report the overexpression and purification of His6-HasA and HasR from S. marcescens in Escherichia coli as well as the preparation and crystallization of the complex holo-His6-HasA–HasR.
2. Materials and methods
2.1. Purification of the holo-His6-HasA–HasR complex
2.1.1. Cloning, cell cultivation and harvesting
His6-HasA (21 500 Da) and HasR (94 800 Da) were cloned and overexpressed in E. coli PAP 105 (Δ [lac pro] thi rpsL supE endA sbcB hsdR F′ [traD36 proAB lacIqZΔM15 Tn10]) and POPc4420 (OmpF− OmpC− LamB−), respectively.
His6-HasA was cloned in a derivative of pQE32 (Qiagen; Létoffé et al., 2005 ▶). In this construct, the following sequence was inserted between the first two N-terminal residues of wild-type HasA: RGSHHHHHHGIRMRARYP. This was confirmed by amino-acid sequencing of the protein. E. coli strain PAP 105(pHis-HasA) was grown in LB medium supplemented with 100 µg ml−1 ampicillin at 310 K to an OD600nm of 0.5. His6-HasA expression was induced with IPTG at 1 mM for 180 min.
HasR was cloned in pFR2 (Létoffé et al., 2004 ▶). Popc4420(pFR2) was grown at 303 K in M9 minimal medium supplemented with 0.2% casaminoacids, 50 µM FeSO4/sodium citrate and 100 µg ml−1 ampicillin. At an OD600nm of 0.5, expression of HasR was induced with 4 × 10−2 mg ml−1 arabinose for 180 min.
The cells were harvested at 5000 rev min−1 for 10 min and resuspended in 10 mM Tris–HCl pH 7.5, 10 mM imidazole for His6-HasA cells and 100 mM Tris–HCl pH 7.5 for HasR cells with protease-inhibitor cocktail (Roche; one tablet per 50 ml). The cells were disrupted in a French press by two successive cycles at 69 MPa; DNAse and RNAse were added at 1 mg ml−1 and incubated with the broken cell suspension for 15 min. Finally, the broken cells were centrifuged at 15 000 rev min−1 for 45 min. The soluble His6-HasA was contained in the supernatant. In the case of HasR, the pellet containing the receptor was resuspended in 100 mM Tris–HCl pH 7.5 and this membrane fraction was kept frozen at 193 K.
2.1.2. Purification of His6-HasA
The supernatant was loaded onto Ni–NTA (Qiagen) packed into a 25 ml column. After a washing step with buffer A (10 mM Tris–HCl pH 7.5, 10 mM imidazole with protease-inhibitor cocktail; Roche, one tablet per 200 ml), His6-HasA was eluted with a step gradient of buffer B (500 mM imidazole in the same buffer). Fractions containing the protein were pooled, concentrated to 5 ml with an ultrafiltration cell (Amicon, exclusion limit 10 000 Da) and dialysed overnight against 10 mM Tris–HCl pH 7.5 containing protease inhibitors. Purification was checked by SDS–PAGE on a 12.5%(w/v) gel.
Part of His6-HasA was already haem-loaded under these culture conditions. To obtain fully haem-loaded protein, His6-HasA was complexed with a solution of haem (bovine haemin, Sigma) using a molar ratio of 1.5 haemin to protein. Haemin was initially dissolved in a minimal volume of 0.1 M NaOH and diluted in the appropriate buffer. Solutions were prepared immediately before use. The haemin concentration was determined from the absorbance at 385 nm using the previously published ∊385 of 58 400 M −1 cm−1 (Dawson et al., 1986 ▶). Holo-His6-HasA was then purified by gel filtration.
2.1.3. Purification of the holo-His6-HasA–HasR complex
All buffers used for the purification of the holo-His6-HasA–HasR complex contained protease inhibitors (one tablet per 200 ml).
The membrane fraction was solubilized with n-tetradecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (CMC 0.014%; ZW3-14, Calbiochem). The membrane fraction from 50 g bacteria was resuspended in 100 ml Tris–HCl pH 7.5, 1 mM MgCl2 and solubilized with 1% ZW3-14 at room temperature for 1 h, followed by centrifugation for 40 min at 20 000 rev min−1 at 277 K. Two successive solubilizations with 2 and 5%(w/v) detergent extracted HasR from the outer membrane. The extracts were pooled to yield solubilized apoHasR.
Holo-His6-HasA was diluted with 50 mM Tris–HCl pH 7.5, 0.08%(w/v) ZW3-14 and loaded in a molar threefold excess compared with estimated amounts of HasR onto a 25 ml nickel–NTA column (Qiagen, 25 ml gel bed volume). Solubilized apoHasR was then applied in order to form the holo-His6-HasA–HasR complex. Previous experiments showed no nonspecific binding between HasR and the column. The complex was eluted with a linear gradient of imidazole (10–500 mM) in the same buffer. The fractions were pooled and concentrated and unliganded holoHasA was finally separated from the complex by gel filtration on Sephacryl S-300 (Amersham, XK 26/70, 350 ml gel bed volume) in 100 mM Tris–HCl pH 7.5, 0.08% ZW3-14. The holo-His6-HasA–HasR complex eluted as a symmetric band at 132 ml, whereas holo-His6-HasA eluted at 172 ml. The fractions containing holo-His6-HasA–HasR were concentrated and detergent-exchanged by ion-exchange chromatography on Q-Sepharose (Amersham, HR 10/10, 8 ml gel bed volume). The zwitterionic detergent was exchanged against various detergents that were investigated during crystallization: N,N-dimethyl dodecylamine oxide (CMC 0.048%; LDAO Fluka), N,N-dimethyldecylamine oxide (CMC 0.095%; DDAO, Fluka), octylpolyoxyethylene 5 octylether (CMC 0.15%; C8E5, Sigma) and polyoxyethylene 4 octylether (CMC 0.22%; C8E4, Bachem). The proteins were washed with ten column volumes of buffer Q (20 mM Tris–HCl pH 7.5 and detergent at four times the CMC of the respective detergent, without protease inhibitor) and the complex was eluted with a linear gradient of 0–1 M NaCl in the same buffer Q.
Finally, the fractions containing holo-His6-HasA–HasR eluting at 0.5 M NaCl were pooled and concentrated to approximately 40 mg ml−1 by ultrafiltration (Vivascience, Vivaspin 6, 50 000 Da). The concentration of NaCl was reduced in two steps of concentration and dilution to a final concentration of around 10−5 M. From a 300 l fermentor, approximately 800 g of wet bacterial pellet was obtained. The final yield was 0.7–0.8 mg of purified complex per gram of HasR-producing bacteria.
2.2. Crystallization and X-ray diffraction
The protein stock solution was diluted with buffer Q to the desired concentration as determined from the absorbance at 277 nm using the theoretical ∊277 of 20 000 M −1 cm−1 plus 140 000 M −1 cm−1 for His6-HasA bound to HasR. Preliminary crystallization screenings using sitting drops were set up in 96-well microplates at various protein concentrations from 5 to 30 mg ml−1 using screening kits (Crystal Screen, Crystal Screen 2, MembFac; Hampton Research). 1 µl protein solution (20 mg ml−1 protein in 20 mM Tris–HCl pH 7.5, 0.6% C8E5) and 1 µl reservoir solution were mixed. The 2 µl drop was equilibrated against 100 µl reservoir. The temperature during crystal growth was set to 291 K. Small crystals appeared after 4–5 weeks with solution No. 27 from Crystal Screen (0.1 M HEPES pH 7.5, 0.2 M sodium citrate, 20% 2-propanol).
Improvement of this initial condition was attempted by the hanging-drop method in 24-well microplates. The final reservoir solution (500 µl) was 0.1 M HEPES pH 7.5, 0.6–0.8 M sodium citrate, 5% 2-propanol. The crystallization drop was formed by mixing 1 µl protein solution (22 mg ml−1 protein in 20 mM Tris–HCl pH 7.5, 0.6% C8E5) with 1 µl reservoir solution. The crystals were grown at 291 K and frozen in liquid nitrogen without cryoprotectant.
Data collection was performed at the Swiss Light Source (SLS) synchrotron, Villigen, Switzerland. Beamline X06SA was equipped with a MAR CCD detector. The wavelength was 1.04 Å. The diffraction was measured at 100 K and data were processed using XDS (Kabsch, 1993 ▶; Diederichs & Karplus, 1997a ▶,b ▶).
3. Results and discussion
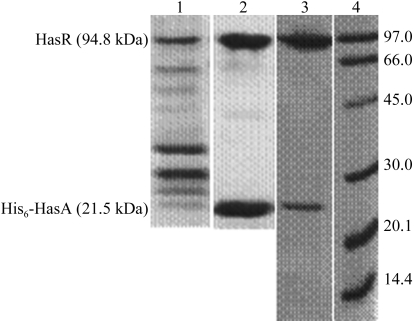
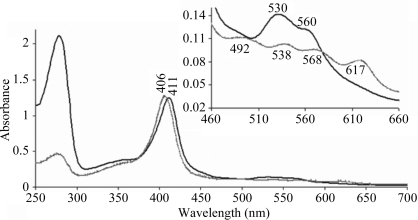
His6-HasA and HasR have been successfully overexpressed in E. coli and purified. His6-HasA behaves like wild-type HasA. It binds haem and the receptor with the same apparent affinities and is able to deliver haem to the bacteria (Létoffé et al., 2005 ▶). Owing to the His6 tag and the affinity between His6-HasA and HasR (K a = 109 M −1), we were able to purify the complex to at least 95% purity as estimated from Coomassie-stained SDS–PAGE (Fig. 1 ▶). An amino-acid analysis revealed a 1:1 stoichiometry between His6-HasA and HasR without any degradation. Haem determination by the pyridine haemochromogen method (De Duve, 1948 ▶) of holo-His6-HasA–HasR shows a 1:1 stoichiometry (one haem bound per protein complex). Spectra of holo-His6-HasA and the holoHis6-HasA–HasR complex taken under oxidative conditions show some differences in the haem-absorption bands: the Soret band is shifted from 406 nm (in the presence of detergents) to 411 nm. In the α/β region, four bands (492, 536, 568 and 618 nm) characterize the haem liganding in holo-His6-HasA. In the complex, the bands at 492 and 618 nm disappear and the bands at 536 and 568 nm shift to 530 and 560 nm, respectively, with an increase in their intensity (Fig. 2 ▶). The spectral differences arise from changes in the haem environment when holoHis6-HasA binds HasR and do not result from haem dissociation from the complex.
Figure 1.
SDS–PAGE gels [12.5%(w/v) acrylamide; coloured by the Coomassie staining technique] showing purification of the holo-His6-HasA–HasR complex. Lane 1, solubilized membrane fraction in 100 mM Tris–HCl pH 7.5, 2%(w/v) ZW3-14; lane 2, complex of holo-His6-HasA (21 500 Da) and HasR (94 800 Da) after the first affinity chromatography step (nickel–NTA) in 50 mM Tris–HCl pH 7.5, 0.08%(w/v) ZW3-14; lane 3, holo-His6-HasA–HasR after the last ion-exchange chromatography step (Q-Sepharose) in 20 mM Tris–HCl pH 7.5, 0.6%(w/v) C8E5; lane 4, marker proteins (kDa; BioRad).
Figure 2.
Absorption spectra of holo-His6-HasA and holo-His6-HasA–HasR. The absorption spectra in the range 250–700 nm under oxidizing conditions of holo-His6-HasA (dotted line) and holo-His6-HasA–HasR (solid line) are shown. The upper insert corresponds to an enlargement of the range 460–700 nm.
Different detergents were tried for crystallization. The type of polar group as well as the length of the hydrophobic chain were varied. N,N-dimethyl dodecylamine oxide (LDAO), N,N-dimethyl decylamine oxide (DDAO), N,N-dimethyl octylamine oxide (ODAO), octylpolyoxyethylene 5 octylether (C8E5) and polyoxyethylene 4 octylether (C8E4) were used. With LDAO, DDAO and ODAO and at various protein concentrations from 5 to 40 mg ml−1 crystals did not grow. Small amphiphiles such as 1,2,3-heptanetriol (Sigma) or C6E5 (Sigma) did not promote crystal growth. In C8E4 or C8E5, only half of the protein concentration is required to observe the same ratio between cleared drops and drops with precipitate compared with LDAO, indicating a lower solubility with these smallest detergents.
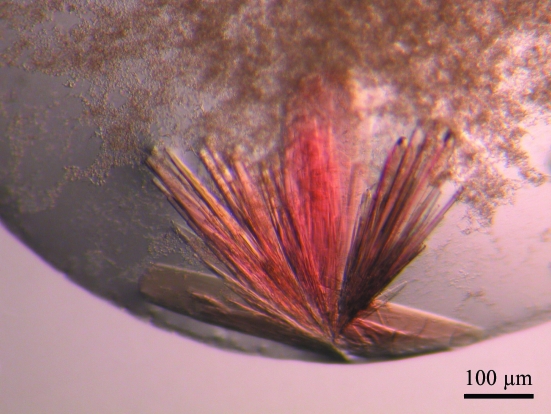
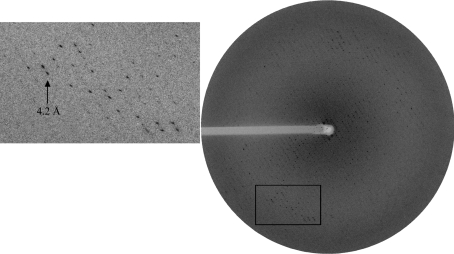
In C8E5, needles and small cubic crystals of 0.010 × 0.010 × 0.010 µm in size grew in 4–5 weeks at 15–24 mg ml−1 protein concentration with condition No. 27 of Crystal Screen using the sitting-drop method. In hanging drops, the crystals obtained at 22 mg ml−1 exhibit a red/brown colour typical of haem. The crystals grew in eight months as plates or long needles of 1000 × 20 × 20–100 µm and are often clustered (Fig. 3 ▶). The largest crystal was frozen in liquid nitrogen. Oscillation photographs were taken at the synchrotron. The reflections were weak and the pattern was anisotropic for resolution higher than 6 Å. Despite this, some reflections were observed to 4.0 Å (Fig. 4 ▶). In vitro, the binding of HasA to HasR is so strong that interaction between both is almost irreversible. Upon storage at 277 K, a solubilized native complex is stable for months. Furthermore, there is no dissociation of the complex even at high salt concentrations. The high stability of the complex as well as the colour and the absorbance of the needles indicate that the crystal consists of the complex holo-His6-HasA–HasR. A native data set was collected to 6.8 Å and data were reduced using the program XDS (Table 1 ▶) (Kabsch, 1993 ▶; Diederichs & Karplus, 1997a ▶,b ▶). The space group is P212121, with unit-cell parameters a = 102.10, b = 182.47, c = 193.98 Å, α = β = γ = 90.00°. The packing density for two complexes (116 300 Da) in the asymmetric unit of the crystal is 3.9 Å3 Da−1, indicating an approximate solvent content of 68% (Matthews, 1968 ▶), which is in agreement with the weak diffraction of our crystal as well as with results typical of membrane-protein crystals.
Figure 3.
Native holo-His6-HasA–HasR crystals grown at the periphery of a hanging drop.
Figure 4.
Oscillation photograph of a holo-His6-HasA–HasR crystal taken under cryogenic conditions.
Table 1. X-ray data collection of the native holo-His6-HasA–HasR crystal.
| Unit-cell parameters (Å, °) | a = 102.10, b = 182.47, c = 193.98, α = β = γ = 90.00 |
| Space group | P212121 |
| Resolution (Å) | 50–6.8 (7.0–6.8) |
| Wavelength (Å) | 1.04 |
| Unique reflections | 6584 (538) |
| Observed reflections | 46083 (3898) |
| Completeness (%) | 99.4 (100) |
| I/σ(I) | 7.87 (4.06) |
| Rmeas† (%) | 26.7 (56.1) |
| Rmrgd-F (%) | 20.5 (35.3) |
We hope that further optimization will yield atomic resolution data. Once determined, the structure of the complex holo-His6-HasA–HasR will represent a sound basis to understand how haem is translocated from HasA to the periplasm through the receptor HasR. Moreover, this insight will be paradigmatic for other haem/haemophore acquisition systems.
Acknowledgments
We thank our colleagues André Schiefner, Mario Mörtl and Kay Diederichs for crystal measurement and data collection at the SLS and for helpful discussions during this work.
References
- Arnoux, P., Haser, R., Izadi, N., Lecroisey, A., Delepierre, M., Wandersman, C. & Czjzek, M. (1999). Nature Struct. Biol.6, 516–520. [DOI] [PubMed] [Google Scholar]
- Biville, F., Cwerman, H., Létoffé, S., Rossi, M. S., Drouet, V., Ghigo, J. M. & Wandersman, C. (2004). Mol. Microbiol.53, 1267–1277. [DOI] [PubMed] [Google Scholar]
- Bracken, C. S., Baer, M. T., Abdur-Rashid, A., Helms, W. & Stojiljkovic, I. (1999). J. Bacteriol.181, 6063–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, M., Endriss, F., Killmann, H. & Braun, V. (2003). J. Bacteriol.185, 5508–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimento, D. P., Mohanty, A. K., Kadner, R. J. & Wiener, M. C. (2003). Nature Struct. Biol.10, 394–401. [DOI] [PubMed] [Google Scholar]
- Dawson, R. M. C., Elliott, D. C., Elliott, W. H. & Jones, K. M. (1986). Data for Biochemical Research, pp. 230–231. Oxford University Press.
- De Duve, C. (1948). Acta Chem. Scand.2, 264–289. [DOI] [PubMed]
- Deniau, C., Gilli, R., Izadi-Pruneyre, N., Létoffé, S., Delepierre, M., Wandersman, C., Briand, C. & Lecroisey, A. (2003). Biochemistry, 42, 10627–10633. [DOI] [PubMed] [Google Scholar]
- Diederichs, K. & Karplus, P. A. (1997a). Nature Struct. Biol.4, 269–275. [DOI] [PubMed] [Google Scholar]
- Diederichs, K. & Karplus, P. A. (1997b). Nature Struct. Biol.4, 592. [DOI] [PubMed]
- Enz, S., Brand, H., Orellana, C., Mahren, S. & Braun, V. (2003a). J. Bacteriol.185, 3745–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enz, S., Brand, H., Orellana, C., Mahren, S. & Braun, V. (2003b). J. Bacteriol.185, 6494. [DOI] [PMC free article] [PubMed]
- Ferguson, A. D., Chakraborty, R., Smith, B. S., Esser, L., van der Helm, D. & Deisenhofer, J. (2002). Science, 295, 1715–1719. [DOI] [PubMed] [Google Scholar]
- Ferguson, A. D., Hofmann, E., Coulton, J. W., Diederichs, K. & Welte, W. (1998). Science, 282, 2215–2220. [DOI] [PubMed] [Google Scholar]
- Ghigo, J. M., Létoffé, S. & Wandersman, C. (1997). J. Bacteriol.179, 3572–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M. W., Muckenthaler, M. U. & Andrews, N. C. (2004). Cell, 117, 285–297. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Létoffé, S., Debarbieux, L., Izadi, N., Delepelaire, P. & Wandersman, C. (2003). Mol. Microbiol.50, 77–88. [DOI] [PubMed] [Google Scholar]
- Létoffé, S., Delepelaire, P. & Wandersman, C. (2004). J. Bacteriol.186, 4067–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létoffé, S., Deniau, C., Wolff, N., Dassa, E., Delepelaire, P., Lecroisey, A. & Wandersman, C. (2001). Mol. Microbiol.41, 439–450. [DOI] [PubMed] [Google Scholar]
- Létoffé, S., Ghigo, J. M. & Wandersman, C. (1994). Proc. Natl Acad. Sci. USA, 91, 9876–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létoffé, S., Nato, F., Goldberg, M. E. & Wandersman, C. (1999). Mol. Microbiol.33, 546–555. [DOI] [PubMed] [Google Scholar]
- Létoffé, S., Wecker, K., Delepierre, M., Delepelaire, P. & Wandersman, C. (2005). J. Bacteriol.187, 4637–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Neilands, J. B. (1995). J. Biol. Chem.270, 26723–26726. [DOI] [PubMed] [Google Scholar]
- Postle, K. & Kadner, R. J. (2003). Mol. Microbiol.49, 869–882. [DOI] [PubMed] [Google Scholar]
- Wandersman, C. & Stojiljkovic, I. (2000). Curr. Opin. Microbiol.3, 215–220. [DOI] [PubMed] [Google Scholar]