The C-terminal domain of the mouse long-chain acyl-CoA thioesterase has been expressed in bacteria and crystallized by vapour diffusion. The crystals diffract to 2.4 Å resolution.
Keywords: mouse long-chain acyl-CoA thioesterase Acot7, 4HBT domain
Abstract
The mammalian long-chain acyl-CoA thioesterase, the enzyme that catalyses the hydrolysis of acyl-CoAs to free fatty acids, contains two fused 4HBT (4-hydroxybenzoyl-CoA thioesterase) motifs. The C-terminal domain of the mouse long-chain acyl-CoA thioesterase (Acot7) has been expressed in bacteria and crystallized. The crystals were obtained by vapour diffusion using PEG 2000 MME as precipitant at pH 7.0 and 290 K. The crystals have the symmetry of space group R32 (unit-cell parameters a = b = 136.83, c = 99.82 Å, γ = 120°). Two molecules are expected in the asymmetric unit. The crystals diffract to 2.4 Å resolution using the laboratory X-ray source and are suitable for crystal structure determination.
1. Introduction
Long-chain acyl-CoAs are intermediates in lipid metabolism and regulators of cellular processes including ion transport, vesicle trafficking, protein phosphorylation and gene expression (Faergeman & Knudsen, 1997 ▶; Hunt & Alexson, 2002 ▶; Yamada, 2005 ▶). Intracellular levels of acyl-CoAs are controlled by the ratio of synthesizing and hydrolysing activities. Acyl-CoA thioesterases (also known as acyl-CoA hydrolases; EC 3.1.2.1 and 3.1.2.2) catalyse the hydrolysis of acyl-CoAs to free fatty acids and CoA-SH. These enzymes are widely distributed among different tissues in mammals (Hunt & Alexson, 2002 ▶).
High levels of long-chain acyl-CoA hydrolysing activity (EC 3.1.2.2) have been detected in mouse and human brain (Anderson & Erwin, 1971 ▶; Yamada, 2005 ▶). Full-length cDNAs corresponding to six different isoforms were cloned from the human brain and shown to be derived from a single gene (Hunt et al., 2005 ▶; Yamada et al., 2002 ▶). When expressed in bacteria, four of the six isoforms showed acyl-CoA hydrolase activity, while two C-terminally truncated isoforms did not (Yamada et al., 2002 ▶). Because several different groups have independently cloned the enzyme, several different names exist, including BACH (Kuramochi et al., 2002 ▶; Yamada et al., 1997 ▶, 1999 ▶), CTE-II (Engberg et al., 1997 ▶) and ACT (Broustas et al., 1996 ▶). The recently suggested revised nomenclature designates this protein as Acot7 (Hunt et al., 2005 ▶). The mouse brain contains a 43 kDa Acot7 as the major isoform and other lesser isoforms including a 50 kDa isoform (Takagi et al., 2004 ▶). The enzyme is specific for acyl-CoA hydrolysis, but has a broad chain-length specificity, hydrolysing acyl-CoAs with carbon numbers C6–C20 (Broustas & Hajra, 1995 ▶; Yamada et al., 1994 ▶, 1996 ▶, 1999 ▶, 2002 ▶).
Acot7 contains two copies of the 4HBT domain (named after 4-hydroxybenzoyl-CoA thioesterase). The structure of 4HBT from Pseudomonas revealed a ‘Hotdog’ fold comprising a β-sheet ‘bun’ wrapped around an α-helical ‘sausage’ (Benning et al., 1998 ▶). This fold was first observed in the structure of Escherichia coli β-hydroxydecanoyl thiol ester dehydratase (FabA; Leesong et al., 1996 ▶) and has since been found in a number of apparently unrelated proteins (Dillon & Bateman, 2004 ▶). The acyl-CoA thioesterases form a distinct class of Hotdog-fold proteins with little available structural information.
To shed light on the molecular function of long-chain acyl-CoA thioesterases, including the enzymatic mechanism and the specificity, we set out to determine the three-dimensional structure of mouse Acot7. As the first step towards this goal, we report here the crystallization and preliminary X-ray diffraction analysis of the C-terminal 4HBT domain of this protein.
2. Experimental methods
2.1. Expression and purification
The DNA encoding amino acids 160–338 of variant 1 of mouse (Mus musculus) Acot7 (GeneInfo Identification No. 19923052; Kuramochi et al., 2002 ▶) was amplified by PCR using cDNA prepared from LPS-stimulated mouse macrophage cells as a template (Wells et al., 2003 ▶). The reactions were catalysed by Triplemaster proofreading, blunt-ended polymerase mix (Eppendorf), using gene-specific primers at a concentration of 10 ng µl−1. The PCR conditions were initial denaturation at 369 K for 4 min, followed by 25 cycles of 369 K for 30 s, 328 K for 30 s and 345 K for 1 min, and finally one cycle of 345 K for 5 min. The PCR product was cloned into the Gateway entry vector pENTR-D-TOPO (Invitrogen) following the manufacturer’s instructions and transformed into chemically competent Top10 cells (Invitrogen) by heat-shock. Colonies were inoculated in 4 ml LB medium in 15 ml Falcon tubes and plasmid DNA was purified from these cultures using a DNA-extraction kit (Roche). Genes cloned into pENTR-D-TOPO were recombined into the expression vector pDEST-17 (Invitrogen) by the Gateway LR reaction following the manufacturer’s instructions; this vector allows expression of a fusion protein containing an N-terminal hexahistidine tag (His tag). The resulting entry and expression vectors were initially assessed by digestion with the restriction enzymes NotI (entry vector) and HindIII and BamHI (expression vector) and confirmed by DNA sequencing.
The expression vector was used to transform the chemically competent E. coli strain BL21(DE3)pLysS by heat-shock. A single colony was inoculated into 20 ml Luria–Bertani (LB) medium and grown overnight at 310 K in the presence of ampicillin and then used to inoculate 2 l LB containing ampicillin. The culture was grown aerobically at 310 K until the OD600nm reached ∼1 and induced with either 1 mM IPTG or 10–30 mM lactose. The temperature was reduced to room temperature upon induction and the culture was grown for a further 12 h. At harvest, the OD600nm was typically ∼5–6 and the yield of recombinant Acot7 was 7–8 mg per litre of culture. The His-tagged protein was purified by immobilized metal-affinity chromatography (IMAC; nickel-affinity gel, Sigma). After elution, the buffer was exchanged using a PD-10 desalting column (BioRad) with 200 mM Tris pH 8.5, 150 mM NaCl, 1 mM DTT and the protein was diluted to ∼3.5 mg ml−1 for crystallizaton. The protein was >95% pure as assessed by SDS–PAGE.
2.2. Crystallization
Prior to crystallization, the protein sample was supplemented with 0.25 mg ml−1 coenzyme A (CoA). Crystallization conditions were screened by the sparse-matrix approach using the hanging-drop vapour-diffusion technique (Jancarik & Kim, 1991 ▶; McPherson, 1982 ▶). 1 µl protein solution was combined with 1 µl reservoir solution and suspended over 0.1 ml reservoir solution. Small crystals were initially observed in 25% polyethylene glycol (PEG) 3350, 0.1 M Tris pH 5.5, 0.2 M ammonium acetate. Crystals could be grown in the pH range 5.5–8, with the best crystals obtained in 20–30% of either PEG 3350, 0.1 M Tris pH 7–8 or 20% PEG 2000 monomethylether (MME), 0.1 M Tris pH 7.0. SDS–PAGE of dissolved crystals confirmed that they were formed by the Acot7 fragment.
2.3. Diffraction data collection
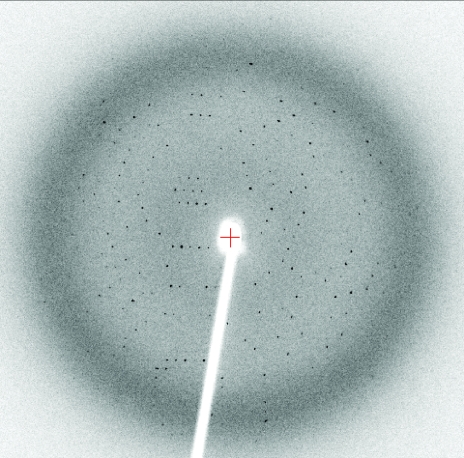
For X-ray diffraction experiments, crystals were transiently soaked in a solution corresponding to the reservoir solution but supplemented with 15% glycerol and were cooled at 100 K in a nitrogen stream (Cryocool, Cryo Industries, New Hampshire, USA). Data were collected from single crystals using an R-AXIS IV++ image-plate detector and Cu Kα radiation from a Rigaku FR-E rotating-anode generator (Rigaku/MSC, Texas, USA). Data were autoindexed and processed with the program CrystalClear (Rigaku/MSC; Table 1 ▶). A representative oscillation image is shown in Fig. 1 ▶.
Table 1. Diffraction data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Reservoir solution | 0.1 M Tris, 20–30% PEG 2000 MME pH 7.0 |
| Space group | R32 |
| Unit-cell parameters | |
| a = b (Å) | 136.83 |
| c (Å) | 99.82 |
| γ (°) | 120 |
| Resolution range (Å) | 30.0–2.4 (2.48–2.40) |
| Observations | 60405 |
| Unique reflections | 14175 |
| Average redundancy | 4.26 (4.19) |
| Completeness (%) | 99.9 (99.6) |
| Rmerge† | 0.05 (0.56) |
| Average I/σ(I) | 12.7 (2.2) |
R
merge = 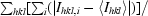
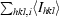 , where I
hkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k and l and 〈I
hkl〉 is the mean intensity of that reflection for I > −3σ(I).
, where I
hkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k and l and 〈I
hkl〉 is the mean intensity of that reflection for I > −3σ(I).
Figure 1.
Oscillation image (1°) of the crystal described in Table 1 ▶. The crystal-to-detector distance is 120 mm; the resolution at the edge of the image is 2.0 Å.
3. Results and discussion
We obtained crystals of the protein corresponding to the C-terminal 4HBT domain of the mouse long-chain acyl-CoA thioesterase (Acot7/mBACH) using either PEG 3350 or PEG 2000 MME as the precipitant. The crystals are rod-shaped (dimensions 0.2 × 0.2 × 0.1 mm) and have the symmetry of space group R32. The crystals appear after a few days and grow to maximum dimensions within three weeks. There are likely to be two molecules of the protein in the asymmetric unit [assuming two molecules, the Matthews coefficient V M (Matthews, 1968 ▶) and the solvent content are 2.0 Å3 Da−1 and 38%, respectively]. The crystals diffract X-rays using the laboratory source to 2.4 Å resolution. The data-collection statistics are shown in Table 1 ▶.
Size-exclusion chromatography suggests the protein expressed from our construct exists in a dimeric form (data not shown), which is consistent with known 4HBT domain structures (Dillon & Bateman, 2004 ▶). Likewise, the 43 kDa isoform showed a molecular weight of 100 kDa using gel filtration (Yamada et al., 1996 ▶, 1999 ▶).
Determination of the crystal structure of this protein will provide important insights into the catalytic mechanism and regulation of the mammalian long-chain acyl-CoA thioesterases. The full-length protein contains two 4HBT domains fused as part of the same polypeptide (the N- and C-terminal domains share 27% sequence identity based on alignment of residues 18–171 and 180–330, respectively); it is currently unknown what the distinct activities of the two domains are and how they cooperate within the full-length proteins. Based on a spectrophotometric assay (Yamada et al., 1994 ▶), our construct of the C-terminal 4HBT domain shows some palmitoyl-CoA hydrolizing activity (J. K. Forwood, W. N. Meng, R. Serek and B. Kobe, unpublished results). While both 4HBT domains are predicted to have the structure corresponding to the Hotdog fold, no structures are currently available of any eukaryotic proteins containing this structural motif.
Acknowledgments
We thank Pawel Listwan for help with cloning and for critically reading the manuscript. This work was supported by the Australian Research Council (ARC; to JLM and BK); BK is a National Health and Medical Research Council of Australia (NHMRC) Research Fellow and ARC Federation Fellow.
References
- Anderson, A. D. & Erwin, V. G. (1971). J. Neurochem.18, 1179–1186. [DOI] [PubMed] [Google Scholar]
- Benning, M. M., Wesenberg, G., Liu, R., Taylor, K. L., Dunaway-Mariano, D. & Holden, H. M. (1998). J. Biol. Chem.273, 33572–33579. [DOI] [PubMed] [Google Scholar]
- Broustas, C. G. & Hajra, A. K. (1995). J. Neurochem.64, 2345–2353. [DOI] [PubMed] [Google Scholar]
- Broustas, C. G., Larkins, L. K., Uhler, M. D. & Hajra, A. K. (1996). J. Biol. Chem.271, 10470–10476. [DOI] [PubMed] [Google Scholar]
- Dillon, S. C. & Bateman, A. (2004). BMC Bioinformatics, 5, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg, S. T., Aoyama, T., Alexson, S. E., Hashimoto, T. & Svensson, L. T. (1997). Biochem. J.323, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman, N. J. & Knudsen, J. (1997). Biochem J.323, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M. C. & Alexson, S. E. (2002). Prog. Lipid Res.41, 99–130. [DOI] [PubMed] [Google Scholar]
- Hunt, M. C., Yamada, J., Maltais, L. J., Wright, M. W., Podesta, E. J. & Alexson, S. E. (2005). J. Lipid Res.46, 2029–2032. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kuramochi, Y., Takagi-Sakuma, M., Kitahara, M., Emori, R., Asaba, Y., Sakaguchi, R., Watanabe, T., Kuroda, J., Hiratsuka, K., Nagae, Y., Suga, T. & Yamada, J. (2002). Brain Res. Mol. Brain Res.98, 81–92. [DOI] [PubMed] [Google Scholar]
- Leesong, M., Henderson, B. S., Gillig, J. R., Schwab, J. M. & Smith, J. L. (1996). Structure, 4, 253–264. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: John Wiley & Sons.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Takagi, M., Kawabe, K., Suga, T. & Yamada, J. (2004). Arch. Biochem. Biophys.429, 100–105. [DOI] [PubMed] [Google Scholar]
- Wells, C. A., Ravasi, T., Faulkner, G. J., Carninci, P., Okazaki, Y., Hayashizaki, Y., Sweet, M., Wainwright, B. J. & Hume, D. A. (2003). BMC Immunol.4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, J. (2005). Amino Acids, 28, 273–278. [DOI] [PubMed] [Google Scholar]
- Yamada, J., Furihata, T., Iida, N., Watanabe, T., Hosokawa, M., Satoh, T., Someya, A., Nagaoka, I. & Suga, T. (1997). Biochem. Biophys. Res. Commun.232, 198–203. [DOI] [PubMed] [Google Scholar]
- Yamada, J., Furihata, T., Tamura, H., Watanabe, T. & Suga, T. (1996). Arch. Biochem. Biophys.326, 106–114. [DOI] [PubMed] [Google Scholar]
- Yamada, J., Kuramochi, Y., Takagi, M., Watanabe, T. & Suga, T. (2002). Biochem. Biophys. Res. Commun.299, 49–56. [DOI] [PubMed] [Google Scholar]
- Yamada, J., Kurata, A., Hirata, M., Taniguchi, T., Takama, H., Furihata, T., Shiratori, K., Iida, N., Takagi-Sakuma, M., Watanabe, T., Kurosaki, K., Endo, T. & Suga, T. (1999). J. Biochem. (Tokyo), 126, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Yamada, J., Matsumoto, I., Furihata, T., Sakuma, M. & Suga, T. (1994). Arch. Biochem. Biophys.308, 118–125. [DOI] [PubMed] [Google Scholar]