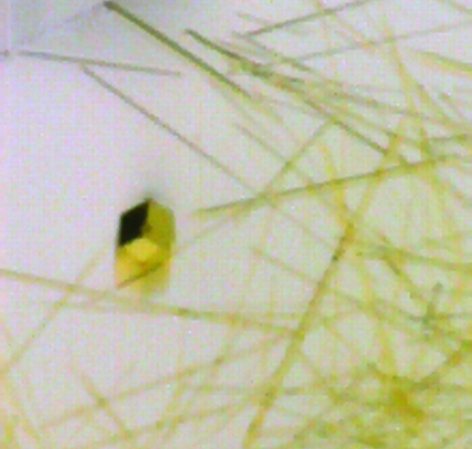
The FAD domain of the NqrF subunit from the Na+-translocating NADH dehydrogenase from V. cholerae has been purified and crystallized. A complete data set was recorded at 3.1 Å.
Keywords: NqrF FAD domain, Na+ translocation, redox pump
Abstract
The Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from pathogenic and marine bacteria is a respiratory complex that couples the exergonic oxidation of NADH by quinone to the transport of Na+ across the membrane. The NqrF subunit oxidizes NADH and transfers the electrons to other redox cofactors in the enzyme. The FAD-containing domain of NqrF has been expressed, purified and crystallized. The purified NqrF FAD domain exhibited high rates of NADH oxidation and contained stoichiometric amounts of the FAD cofactor. Initial crystallization of the flavin domain was achieved by the sitting-drop technique using a Cartesian MicroSys4000 robot. Optimization of the crystallization conditions yielded yellow hexagonal crystals with dimensions of 30 × 30 × 70 µm. The protein mainly crystallizes in long hexagonal needles with a diameter of up to 30 µm. Crystals diffract to 2.8 Å and belong to space group P622, with unit-cell parameters a = b = 145.3, c = 90.2 Å, α = β = 90, γ = 120°.
1. Introduction
Respiratory NADH dehydrogenases couple the exergonic reduction of quinone to the uphill transport of protons or Na+ across energy-conserving membranes. In mitochondria, NADH is oxidized by complex I, a multisubunit redox pump composed of 46 subunits (Carroll et al., 2005 ▶). In some bacteria, such as the human pathogen Vibrio cholerae, the primary respiratory complex is the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR), which consists of six subunits, NqrA–F, and contains one Fe–S centre, two covalently bound FMNs, one non-covalently bound FAD, one riboflavin and ubiquinone-8 as prosthetic groups (Bogachev & Verkhovsky, 2005 ▶; Hayashi & Unemoto, 2004 ▶; Türk et al., 2004 ▶). Electrogenic NADH:quinone oxidoreductases are of the utmost importance for energy conversion in cells. In humans, dysfunction of the mitochondrial complex I represents a frequently encountered inherited defect of the oxidative phosphorylation system (Vogel et al., 2004 ▶). In V. cholerae, Na+-NQR maintains an electrochemical Na+ gradient across the inner bacterial membrane which strongly influences the production of virulence factors (Häse & Mekalanos, 1999 ▶). Therefore, Na+-NQR represents a putative target for novel antibiotics. So far, no structural information at atomic resolution is available for respiratory NADH dehydrogenases. With only six subunits, Na+-NQR is ideally suited for study of the structure and function of cation-translocating NADH:quinone oxidoreductases. The initial oxidation of NADH by the Na+-NQR complex is catalyzed by the membrane-bound NqrF subunit, which displays a clearly defined domain structure. While the N-terminal domain of NqrF harbours a [2Fe–2S] cluster, binding sites for the non-covalently bound FAD and NADH are located in the C-terminal domain. This C-terminal part of NqrF, which was overproduced as a soluble protein in V. cholerae, catalyzed NADH oxidation and contained FAD in stoichiometric amounts (Türk et al., 2004 ▶). Here, we report an optimized purification protocol for the FAD domain of the NqrF subunit, its crystallization and preliminary X-ray analysis.
2. Material and methods
2.1. Protein expression and purification
The FAD domain of NqrF was purified as described previously (Türk et al., 2004 ▶) with minor modifications. The purification includes affinity chromatography utilizing an N-terminal hexahistidine tag and ion-exchange chromatography. Briefly, the NqrF FAD domain (amino acids 129–408) was cloned into pEC422 and transformed into V. cholerae O395 N1. V. cholerae O395 N1 was grown in LB medium at 310 K and expression was induced by the addition of 10 mM arabinose. After 4 h induction, the cells were harvested by centrifugation. The cells were suspended in 10 mM Tris–HCl pH 7.4, 0.3 M NaCl, 0.1 mM diisopropylfluorophosphate, 1.0 mM dithiothreitol, 5 mM MgCl2 and traces of DNase I. The cells were broken by French press treatment and the crude extracts were centrifuged for 1 h at 100 000g. The supernatant was applied onto an Ni–NTA column (Quiagen) equilibrated in 20 mM Tris–HCl pH 8.0, 0.5 M NaCl (buffer A). The column was washed with buffer A containing 30 mM imidazole. The NqrF FAD domain was eluted with buffer A containing 400 mM imidazole. The N-terminal polyhistidine tag was removed by thrombin proteolysis. Prior to proteolysis, the buffer was exchanged to 50 mM Tris–HCl pH 7.5, 0.15 M NaCl, the protein concentration was adjusted to approximately 1 mg ml−1 by ultrafiltration (Amicon, 10 kDa cutoff) and CaCl2 was added to a final concentration of 2.5 mM. 10 U thrombin (GE Healthcare) per milligram of protein was allowed to react with the protein for 2 h at 298 K. The digest was again passed over an Ni–NTA column in order to remove undigested protein as well as the histidine tag. The flowthrough was diluted tenfold with 50 mM HEPES–NaOH pH 7.0 and applied onto a Mono Q HR10/10 column (GE Healthcare) connected to an Äkta purifier system (GE Healthcare). The NqrF FAD domain was eluted in a linear gradient of 0–0.4 M NaCl.
2.2. Protein crystallization and preliminary X-ray analysis
Prior to crystallization, the buffer was exchanged to 5 mM Tris–HCl pH 7.5 and the protein concentration was adjusted to 7 mg ml−1. Crystallization conditions were tested by robotic screening on the nanolitre scale using a nanodrop crystallization robot (Cartesian technologies). Each drop was prepared by mixing 100 nl protein solution with an equal amount of crystallization solution and was equilibrated by vapour diffusion against 100 µl crystallization solution in the reservoir at 293 K. Crystals for X-ray analysis were grown using the hanging-drop vapour-diffusion method by mixing 2 µl protein solution with 2 µl reservoir solution [0.1 M MES–NaOH pH 6.6, 2.2 M (NH4)2SO4, 0.6% PEG 400] equilibrated against 500 µl reservoir solution at 293 K. The crystals were soaked for 2 min in the crystallization solution containing 30%(v/v) glycerol as cryoprotectant and were immediately flash-frozen in the cryo-nitrogen stream. X-ray data were collected at 100 K using monochromatic synchrotron radiation (λ = 0.9787 Å) at the beamline X06SA, Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland. The diffraction data were processed with the program package XDS (Kabsch, 1988 ▶).
3. Results and discussion
Trials to express the NqrF FAD domain using pET vectors in Escherichia coli were not successful. It was most likely that the FAD domain does not fold properly in E. coli and is proteolytically degraded. Such misfolding might be caused by the incomplete incorporation of FAD or the absence of chaperones specific for V. cholerae. Therefore, the expression was performed in a V. cholerae strain using a pBR322-derived vector. The purification yield from 14 g of cells (wet weight) was approximately 9 mg of pure NqrF FAD domain still containing the His tag. Attempts to crystallize this protein failed. We concluded that the His tag prevented crystallization. The N-terminal His tag and the linker region comprise 20 amino acids which presumably do not adopt a stable conformation. Therefore, we removed the His tag and included a further protein-purification step on a high-resolution MonoQ column. The yield of highly pure NqrF FAD domain devoid of the His tag was 3 mg from 14 g wet weight of cells. The cleavage of the His tag was 95% complete as judged by MALDI–MS. Owing to the limited amounts of protein, we performed crystallization screening with a Cartesian robotic system using drop volumes of 100 nl. In such screens needle-like crystals of the NqrF FAD domain were obtained with several solutions containing (NH4)2SO4 as precipitant. These needles had a maximum diameter of 5 µm and diffracted to 12 Å. The crystallization conditions were refined, varying the pH and the (NH4)2SO4 concentration and adding further precipitants such as PEG. In optimized crystallization trials NqrF FAD domain crystals were grown using the hanging-drop vapour-diffusion technique in 2 µl drops within two months (Fig. 1 ▶). However, most of the protein still formed thin needles that were not suitable for X-ray analysis. Some crystals reached dimensions of 30 × 30 × 70 µm. Using synchrotron radiation, these crystals diffracted to 2.8 Å and a complete data set was recorded at 3.1 Å resolution. Data statistics are given in Table 1 ▶. The space group was determined to be P622, with unit-cell parameters a = b = 145.3, c = 90.2 Å, α = β = 90, γ = 120°. The unit-cell parameters indicated the presence of two molecules in the asymmetric unit. In solution, the FAD domain was monomeric as determined by size-exclusion chromatography. Based on a molecular weight of 32.3 kDa, the solvent content of the crystals was calculated to be 44%, with a Matthews coefficient of 2.2 Å3 Da−1. Ag+ is a strong specific inhibitor of NqrF (Steuber et al., 1997 ▶). We are currently trying to obtain an Ag+ heavy-atom derivative for phase determination. The identification of the Ag+-binding site in the NqrF FAD domain will elucidate the molecular mechanism of Ag+ inhibition. We are also searching for conditions allowing protein expression on minimal medium in V. cholerae, which is a prerequisite for the production of SeMet derivatives and phase determination by MAD.
Figure 1.
Crystals of the NqrF FAD domain. Along with long thin needles, larger hexagonal crystals grew within two months. The dimensions of the hexagonal crystal are 30 × 30 × 70 µm.
Table 1. X-ray data statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P622 |
| Unit-cell parameters (Å, °) | a = b = 145.3, c = 90.2, α = β = 90, γ = 120 |
| Resolution (Å) | 50–3.1 (3.4–3.1) |
| Wavelength (Å) | 0.9787 |
| Completeness (%) | 100.0 (99.9) |
| No. of unique reflections | 10657 (2493) |
| Rmerge (%) | 7.8 (22.5) |
| I/σ(I) | 21.4 (6.2) |
| No. of molecules per ASU | 2 |
| Matthews coefficient (Å3 Da−1) | 2.2 |
| Solvent content (%) | 44.1 |
Acknowledgments
We thank Beat Blattmann and Po-Chi Lin, University of Zurich for setting up the initial crystallization screens. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to GF, TR-SFB11), Swiss National Science Foundation (to JS), Vontobel Stiftung (to JS) and Roche Research Foundation (to KT).
References
- Bogachev, A. V. & Verkhovsky, M. I. (2005). Biochemistry (Mosc.), 70, 143–149. [DOI] [PubMed] [Google Scholar]
- Carroll, J., Fearnley, I. M., Skehel, J. M., Runswick, M. J., Shannon, R. J., Hirst, J. & Walker, J. E. (2005). Mol. Cell. Proteomics, 4, 693–699. [DOI] [PubMed] [Google Scholar]
- Häse, C. C. & Mekalanos, J. J. (1999). Proc. Natl Acad. Sci. USA, 96, 3183–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M. & Unemoto, T. (2004). Respiration in Archaea and Bacteria: Diversity of Prokaryotic Electron-Transport Carriers, edited by D. Zannoni, Vol. 15, pp. 15–174. Dordrecht: Kluwer Academic Publishers.
- Kabsch, W. (1988). J Appl. Cryst.21, 67–71. [Google Scholar]
- Steuber, J., Krebs, W. & Dimroth, P. (1997). Eur. J. Biochem.249, 770–776. [DOI] [PubMed] [Google Scholar]
- Türk, K., Puhar, A., Neese, F., Bill, E., Fritz, G. & Steuber, J. (2004). J. Biol. Chem.279, 21349–21355. [DOI] [PubMed] [Google Scholar]
- Vogel, R., Nijtmans, L., Ugalde, C., van den Heuvel, L. & Smeitink, J. (2004). Curr. Opin. Neurol.17, 179–186. [DOI] [PubMed] [Google Scholar]