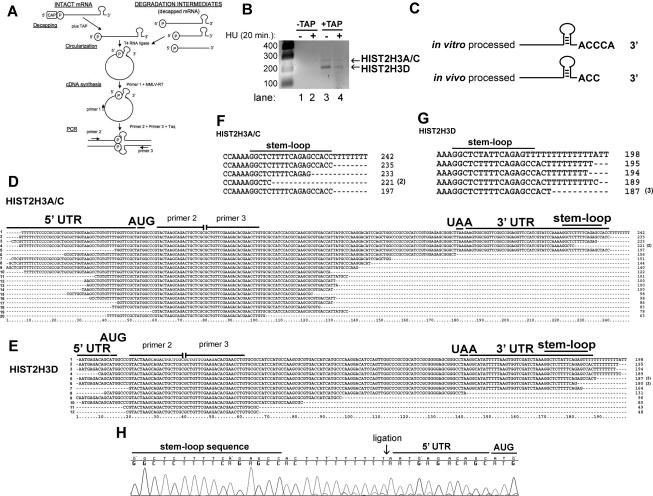
Figure 4.
Detection of the 5′ and 3′ ends of capped histone H3 mRNA in vivo and determination of the sequence of decapped histone H3 mRNA degradation intermediates. (A) The cRT–PCR strategy to determine the ends of intact histone mRNA and degradation intermediates is shown (adapted from Couttet et al. 1997; © 1997 National Academy of Sciences, USA). (B) cRT–PCR reactions with and without decapping from total cell RNA were separated on 1.5% agarose gels, and amplicons were detected by ethidium bromide staining. Two human histone H3 mRNAs are visualized in the +TAP lanes. (C) The 3′ end of a synthetic histone pre-mRNA substrate processed in vitro and decapped differs from histone mRNA decapped and retrieved from the cell. In vitro processed pre-mRNA and total cell RNA were decapped (+TAP), subjected to cRT–PCR, and cloned. Complete sequences of these experiments are presented in Supplemental Figure S4. (D,E) cRT–PCR was performed on total cell RNA, and the histone H3 products were amplified, cloned, and sequenced. HIST2H3A/C (D) or HIST2H3D (E) mRNAs that were not capped were cloned by cRT–PCR. Primers 2 and 3 represent sites where we targeted amplification toward the 5′ and 3′ ends. ORF sequences between these two oligonucleotides are missing. Numbers in parentheses are the number of times each clone was obtained. (F,G) The 3′ ends from the largest HIST2H3A/C (F) and HIST2H3D (G) mRNA degradation intermediates are shown. The number of times each sequence was obtained is indicated by a number in parentheses. (H) Chromatogram from a HIST2H3D mRNA degradation intermediate containing eight untemplated Us (Ts in the DNA sequence). The 3′ and 5′ ends were ligated (arrow) together.