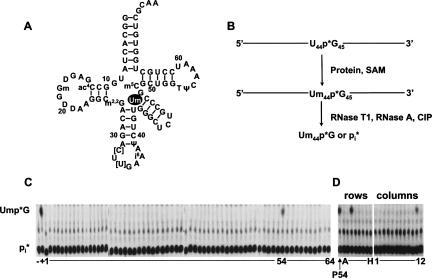
FIGURE 1.
Identification of yeast ORF associated with Um44 methyltransferase activity of tRNASer(UGA). (A) Two-dimensional structure of S. cerevisiae tRNASer(UGA). tRNA is numbered according to convention, in this case with no residue 17, an additional residue 20a, and an 11 residue variable arm between Um44 and G48. [C], [U], uncharacterized C and U modifications. Um44 is shown in white font within a black oval. (B) Assay scheme to detect 2′-O-methylation of tRNASer uniquely labeled at U44 (U44*Ser). Treatment with RNaseT1, RNaseA and calf intestinal phosphatase (CIP) yields Ump*G if U44 is methylated and inorganic phosphate (Pi) if the substrate is unmodified. (C) Assay of a genomic collection of GST-ORF pools for Um44 methyltransferase activity. GST-ORF fusion proteins were purified and assayed for tRNASer Um44 2′-O-methyltransferase using 2 μL of each GST-ORF fusion protein pool, 0.5 mM S-adenosyl methionine, and ∼1 nM U44*Ser tRNA in methyltransferase buffer at 30°C for 12 h, and then RNA was digested with RNase T1, RNase A, and CIP to produce Ump*G dinucleotide or inorganic phosphate, which were resolved by thin layer chromatography as described in Materials and Methods. (Lane −) no protein; (lane +), yeast crude extract. (D) Deconvolution of subpools of Plate 54 to identify the ORF associated with Um44 methyltransferase. (P54) GST-ORF proteins purified from pool 54.