Abstract
Despite the central role of group II introns in eukaryotic gene expression and their importance as biophysical and evolutionary model systems, group II intron tertiary structure is not well understood. In order to characterize the architectural organization of intron ai5γ, we incorporated the photoreactive nucleotides s4U and s6dG at specific locations within the intron core and monitored the formation of cross-links in folded complexes. The resulting data reveal the locations for many of the most conserved, catalytically important regions of the intron (i.e., the J2/3 linker region, the IC1(i-ii) bulge in domain 1, the bulge of D5, and the 5′-splice site), showing that all of these elements are closely colocalized. In addition, we show by nucleotide analog interference mapping (NAIM) that a specific functional group in J2/3 plays a role in first-step catalysis, which is consistent with its apparent proximity to other first-step components. These results extend our understanding of active-site architecture during the first step of group II intron self-splicing and they provide a structural basis for spliceosomal comparison.
Keywords: RNA structure, RNA folding, catalysis, ribozyme, splicing
INTRODUCTION
Self-splicing group II introns are a class of genetic elements that are commonly found within intergenic regions of prokaryotes as well as in the organelles of yeast, algae, and plants (Bonen and Vogel 2001; Lehmann and Schmidt 2003). These elements often reside within critical genes of these organisms, so that their splicing is essential for host viability (Lehmann and Schmidt 2003; Pyle and Lambowitz 2006). Group II introns can also reverse-splice, allowing them to behave as mobile genetic elements (Pyle and Lambowitz 2006). Phylogenetically, group II introns can be subdivided into three subfamilies, IIA, IIB, and IIC, although the majority of biochemical and structural studies have focused on the IIB class (Toor et al. 2001; Toro 2003). Here, we focus on the ai5γ group IIB intron from Saccharomyces cerevisae, which has been extensively characterized by genetics, biochemistry, and biophysical analysis (Lehmann and Schmidt 2003; Pyle and Lambowitz 2006).
All group II introns share a common secondary structural organization of six domains (D1–D6) as well as a common mechanism characterized by two consecutive transesterification reactions that result in intron removal and exon ligation (Michel et al. 1989; Pyle and Lambowitz 2006). Domains 1 (D1) and 5 (D5) are the only elements that are absolutely essential for minimal catalytic activity of the intron (Koch et al. 1992; Michels and Pyle 1995). Domain 6 (D6) is necessary for the branching pathway, whereby a bulged adenosine is utilized as the nucleophile that attacks the 5′-splice site (Lehmann and Schmidt 2003). However, group II introns can also undergo an alternative hydrolysis pathway, both in vivo and in vitro, in which a water molecule serves as the nucleophile during the first step of splicing (Peebles et al. 1987; Jarrell et al. 1988b; Daniels et al. 1996; Podar et al. 1998a).
Despite the importance of group II introns in biology (Pyle and Lambowitz 2006), information on their three-dimensional structures is limited. A lack of sequence conservation and phylogenetic covariations have hindered attempts to predict major architectural features of the intron. Furthermore, in the absence of high-resolution structural information, the spatial organization of critical active-site regions and their role in catalysis has remained difficult to understand. Despite these challenges, numerous studies have defined the approximate position and function of specific intronic regions, such as the highly conserved domain 5 (D5) (for reviews, see Lehmann and Schmidt 2003; Pyle and Lambowitz 2006). The relative spatial orientation of these motifs has been established and three-dimensional models of group II introns have been constructed (Costa et al. 2000; Noah and Lambowitz 2003; de Lencastre et al. 2005). These efforts were greatly facilitated by the identification of docking receptors for the group II intron branch site and the 3′-splice site (de Lencastre et al. 2005; Hamill and Pyle 2006). However, similarly detailed information about 5′-splice site orientation has been lacking.
Intronic nucleotides immediately downstream from the 5′-splice site are highly conserved and have been demonstrated by genetic and chemogenetic methods to be essential for splice-site selection and catalytic function (Michel et al. 1989; Jacquier and Michel 1990; Chanfreau and Jacquier 1994; Boudvillain and Pyle 1998; Boudvillain et al. 2000). Nucleotide analog interference mapping (NAIM) and nucleotide analog interference suppression (NAIS) have shown that several of these residues are key active-site components (Boudvillain and Pyle 1998; Boudvillain et al. 2000). In many cases, every nucleotide functional group, including phosphate moieties, the sugar backbone, as well as atoms in both the minor and the major grooves, contribute to reactivity (Boudvillain and Pyle 1998). These findings indicate that the 5′ terminus of the intron participates in a complex network of interactions that mediate both steps of splicing.
Nonetheless, there is limited information about the tertiary contacts that are made by these nucleotides. The first nucleotide of the intron (G1) has been proposed to pair with the penultimate nucleotide of the intron (Chanfreau and Jacquier 1993). However, this interaction has only been proposed to occur during the second step of splicing; the role of G1 during the first step has remained elusive (Chanfreau and Jacquier 1993). Similarly, the third and fourth nucleotides of the intron have been shown to form a tandem set of Watson–Crick pairing interactions with other nucleotides in D1 (ε–ε′) (Michel et al. 1989; Jacquier and Michel 1990). However, the clear importance of additional functional groups on these nucleotides suggests that they participate in additional contacts as part of a more elaborate substructure (Jacquier and Michel 1990; Boudvillain and Pyle 1998).
Many studies have demonstrated the central importance of nucleotides in D5 (Chanfreau and Jacquier 1994; Peebles et al. 1995; Abramovitz et al. 1996; Boudvillain et al. 2000). A remaining mystery, however, is the role of functionalities in the essential 2-nucleotide (nt) bulge that separates the upper and lower stems of the D5 hairpin. Although only semiconserved (5′Ay), the bulge cannot be deleted without complete loss of catalytic activity, and backbone functional groups in this region are essential for function (Chanfreau and Jacquier 1994; Abramovitz et al. 1996; Schmidt et al. 1996; Boudvillain and Pyle 1998). For instance, a single sulfur substitution of the nonbridging Rp oxygen on C839 in ai5γ eliminates all chemical reactivity (Chanfreau and Jacquier 1994; Boudvillain and Pyle 1998). Importantly, the bulge has been strongly implicated in metal binding (Sigel et al. 2000; Gordon and Piccirilli 2001; Seetharaman et al. 2006), which is particularly interesting given that a similar bulge structure within the spliceosomal U2/U6 pairing also binds metals that are required for function (Yean et al. 2000). Despite its clear importance, the local conformation and tertiary structural arrangement of the D5 bulge region is not well understood. Crystal and NMR structures of D5 display markedly different conformations and the spatial location of the bulge within the group II core is unclear (Zhang and Doudna 2002; Sigel et al. 2004; Seetharaman et al. 2006). In addition, recent NMR studies of D5 in complex with D123 indicate that D5 bulge nucleotides change conformation upon D123 binding, suggesting that the organization of the bulge nucleotides may remain disordered until binding to a docking site (Gumbs et al. 2006; Seetharaman et al. 2006). It is, therefore, of critical importance to ascertain structural information about the tertiary location of these nucleotides in the context of the catalytically competent ribozyme.
In this study, we present a network of site-directed cross-links that reveal the proximity of three catalytically essential and conserved regions of the group II intron: the 5′ terminus of D1, the bulge of D5, and J2/3. The specific pattern of these photo-cross-links allows us to predict the spatial arrangement of these regions with respect to each other and to other core structural elements. In a complementary analysis, we use NAIM to identify critical atoms for the function of J2/3, thereby implicating the N7 group of G588 as specifically important for the first step of splicing. These results provide direct evidence that conserved and catalytically important residues in the D5 bulge and in J2/3 are proximal to the 5′-splice site, suggesting that they play a direct role in active-site assembly and/or catalysis.
RESULTS
Cross-links from the 5′ end of Domain 1
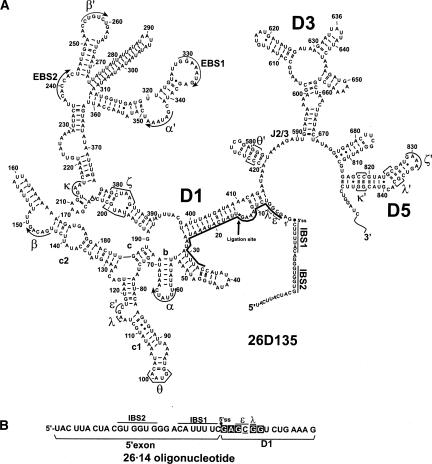
To investigate the tertiary organization of active-site constituents near the 5′-splice site, a group II intron construct was engineered to allow the incorporation of single-atom modifications at the 5′ terminus of the intron (Fig. 1). Using splinted ligation (Moore and Sharp 1992), a synthetic oligonucleotide that consists of a short 5′ exon and the first 14 nt of the intron (26·14) was joined to a truncated D135 transcript (D135-14). The resulting 611-nt molecule (26D135) contains all of the nucleotides that are important for the first step of splicing, such as a 5′exon that bears IBS1 and IBS2, an intact Domain 1, intact junction regions, a hairpin replacement of D2 that preserves the θ–θ′ interaction, as well as Domains 3 and 5 (Fig. 1). When incubated in reaction buffer (0.5M KCl, 0.1M MgCl2 at pH 7.5, at 42°C), 26D135 releases its 5′ exon, thereby undergoing the first step of splicing through hydrolysis (Podar et al. 1995b). To ensure that the synthesis and ligation steps did not damage the molecule, reactivity of a ligated 26D135 was compared with that of a contiguous transcript of the same sequence and length. Activity was found to be unaffected by the semisynthetic procedures (kobs for transcribed and ligated 26D135 were (6.8 ± 0.2) × 10−2 min−1 and (5.8 ± 0.3) × 10−2 min−1, respectively; amplitudes (i.e., reaction extents) for transcribed and ligated 26D123 were 94% (±3.2) and 88% (±1.0), respectively.
FIGURE 1.
Incorporation of photo cross-linkable analogs near the 5′-splice site of a group II intron ribozyme. (A) A group II intron ribozyme bearing domains 1, 3, and 5 (D135), but lacking the first 14 nt of the intron, was ligated via a DNA splint (thick black line) to a synthetic oligonucleotide (26·14) containing 26 nt of the 5′ exon and 14 nt of D1 to reconstitute a semisynthetic 26D135 ribozyme. Ligation site is indicated by an asterisk (*). (B) Sequence of 26·14 oligonucleotides containing site-specifically incorporated cross-linkable analogs s6dG (boxes) and s4U (underlined) at the indicated positions.
This semisynthetic scheme was utilized to prepare a large family of photoactivatable constructs for probing the tertiary structure near the 5′-splice site (5′ss). Six different 26·14 oligonucleotides were synthesized, each bearing the short-range cross-linkable analog 4-thiouridine (s4U) or 6-thiodeoxyguanosine (s6dG) at a specific, single-site location (Fig. 1). Ligation of these oligonucleotides to D135-14 allowed us to probe the spatial location of intron nucleotides 1–6. To ensure that the modified nucleotides did not intrinsically impair reactivity of the intron, all ligated molecules were tested for their ability to complete the first step of splicing (data not shown). We observed that all substituted molecules were reactive. One substitution (C4:s4U4) caused a reduction in the rate constant for the first-step hydrolysis reaction (data not shown), which may reflect a loss of stability for the predicted base pair between C4 and G116 (Jacquier and Michel 1990), or it may reflect the loss of an unidentified tertiary interaction.
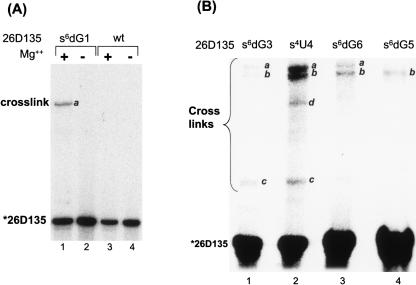
Cross-link formation is Mg++ dependent
When analyzed by PAGE, it was clear that UV-irradiated 26D135 molecules form several species with an electrophoretic mobility that is significantly slower than the 5′-labeled starting material (Fig. 2). The low-mobility product species were observed only when a cross-linkable analog was incorporated into 26D135 and only in the presence of Mg++ (Fig. 2A), which is essential for proper folding of the molecule (Qin and Pyle 1997; Swisher et al. 2002). Although the pattern and intensity of observed cross-links depended on the location of the cross-linkable analog, the cross-linked species could be classified into three major groups—cross-links a, b, and c—according to their electrophoretic mobility on denaturing PAGE (Fig. 2). An additional cross-link (Fig. 2B, cross-link d), which was observed with only one of the analogs, could not be mapped despite numerous attempts and, therefore, was not further characterized. Using primers complementary to different parts of the intron and RT primer extension analysis, the locations of all remaining cross-links were identified.
FIGURE 2.
Cross-links from the 5′ end of D1. (A) A 5′ 32P-labeled 26D135 containing a s6dG analog at the first nucleotide of the intron forms a cross-link upon exposure to UV light (366 nm) after preincubation in reaction conditions (0.5 M KCl, 0.1 M MgCl2, 42°C, 15 min) (lane 1). The cross-link is dependent on Mg++ (lane 2), as well as on the presence of the cross-linking analog (lanes 3,4). (B) Cross-links formed by 26D135 containing s6dG3, s4U4, s6dG6, or s6dG5. Cross-links are annotated (a, b, and c) according to their relative electrophoretic mobility and were subsequently mapped by reverse transcription. Band d could not be mapped and was not characterized further.
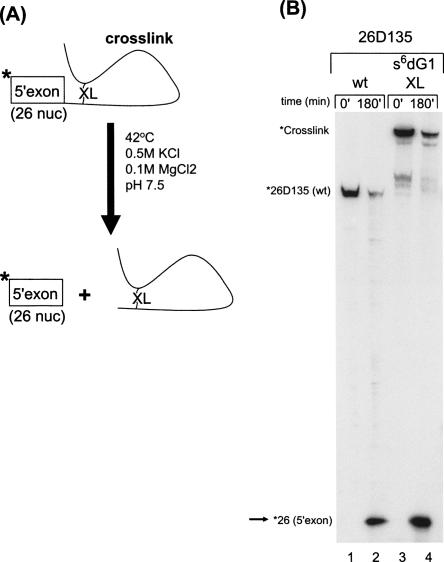
The magnesium dependence of cross-link formation is consistent with proper folding of the intron core structure. However, catalytic reactivity of the cross-linked species would provide further evidence that they represent biologically relevant forms of the ai5γ intron. To this end, cross-linked RNAs were suspended in reaction buffer and the release of 5′-exon was monitored (Fig. 3). We observe that the cross-linked species are capable of undergoing the first step of splicing (Fig. 3B; data not shown), which indicates that the cross-links do not interfere with catalytic function, and that the cross-linked RNA represents a properly folded form of the intron.
FIGURE 3.
Activity of the cross-linked species. (A) The scheme for testing cross-link activity. Isolated cross-linked material (denoted by “XL”) was incubated in standard reaction conditions (0.5 M KCl, 0.1 M MgCl2, 42°C), and the release of 5′ end-labeled 5′ exon was monitored. (B) Gel showing the appearance of the 5′ 32P-labeled 26-nt 5′ exon (indicated by arrow, *26[5′-exon]) when an s6dG1 cross-link is incubated under hydrolysis reaction conditions (lanes 3,4). The minor bands in lanes 3 and 4 represent copurifying cross-links. A 5′ end-labeled wt 26D135 control shows similar reactivity (lanes 1,2).
Mapping of cross-links
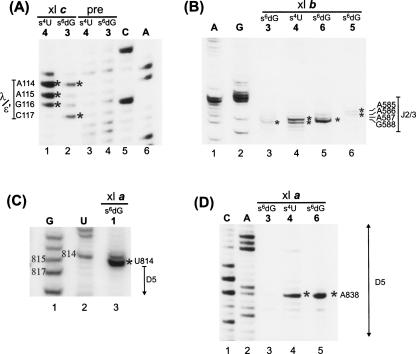
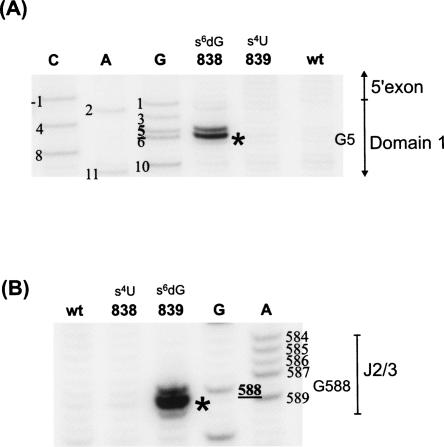
Sites of cross-linking between photoreactive nucleotides in 26D135 and other regions of the intron were mapped by reverse-transcription (Fig. 4; Table 1). This analysis indicates that the 5′ terminus of the intron is in close proximity to three major areas of the intron—the ε′/λ loop of the c1 stem in D1, highly conserved nucleotides in J2/3, and specific regions of D5. All three regions have been previously implicated in catalysis, which is consistent with their close proximity in three-dimensional space.
FIGURE 4.
Mapping of cross-links by reverse transcription primer extension. Cross-links are indicated by an asterisk (*). They were identified by the appearance of an RT stop 1 nt downstream from the corresponding position in a sequencing lane (see Materials and Methods). A, G, C, and U indicate dideoxy sequencing lanes. Nonirradiated precursor “pre” was used as a control to detect natural RT stops. Cross-linked species (i.e., a, b, and c) are designated as in Figure 2. (A) Cross-link c from s4U4 (lane 1) and s6dG3 (lane 2) to the ε′/λ region in D1. Precursor (pre) molecules of s6dG3 and s4U4 (lanes 3,4) are the non-UV irradiated controls for natural RT stops. (B) Mapping of cross-link b identifies cross-links from s6dG3, s4U4, s6dG6, and s6dG5 to the J2/3 region of the intron (note: lane 3 has been contrast enhanced to show band). (C) Cross-link a from s6dG1 to U814 near D5. (D) Cross-link a from s4U4 and s6dG6 to A838 in the bulge of D5.
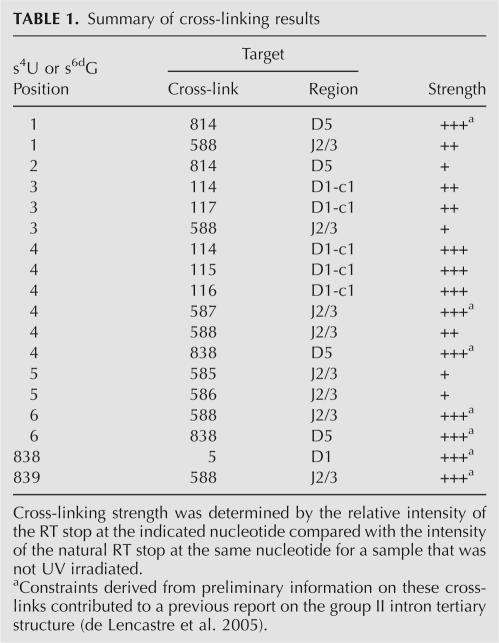
TABLE 1.
Summary of cross-linking results
The first major cluster of cross-links involves an asymmetric loop in D1c1 that spans nucleotides 114–117. This region harbors nucleotides that have been proposed, on the basis of phylogenetic and biochemical evidence, to form the ε–ε′ interaction (Jacquier and Michel 1990) and the λ–λ′ interaction (Boudvillain et al. 2000). It also contains nucleotide A114, which is highly conserved but has an undetermined function (Michel et al. 1989). When nucleotide C4 is substituted with s4U (s4U4), it forms cross-links with A114, A115, and G116, whereas an s6dG substitution at G3 (s6dG3) forms cross-links with A114 and C117 (Fig. 4). No other cross-links are found to cause RT stops in this region (data not shown), indicating that the cluster of cross-links is specific to cross-link c from s6dG3 and s4U4. The observation of cross-links from nucleotides 3 and 4 to nucleotides in D1c1, together with the striking polarity of their formation, is consistent with the proposed ε–ε′ interaction involving the C4-G116 and G3-C117 base pairs (Jacquier and Michel 1990), and it represents direct structural evidence that these nucleotides are indeed proximal in space.
The second major target of cross-linking from photoactivated nucleotides at the 5′-splice site is the linker between domains 2 and 3 (J2/3), which contains two highly conserved nucleotides, A587 and G588, which are important for the catalysis of splicing (Jacquier and Michel 1990; Ho Faix 1998; Mikheeva et al. 2000). Specifically, cross-links in set b from s6dG3, s4U4, s6dG5, and s6dG6 were mapped to the J2/3 region (Fig. 4B). The s6dG3 forms a weak cross-link to G588 (Fig. 4B, lane 3), s6dG5 cross-links weakly to A586 (Fig. 4B, lane 6), and s6dG6 forms a strong cross-link to G588 (Fig. 4B, lane 5). In addition, s4U4 is found to cross-link strongly to A587 and moderately to G588 (Fig. 4B, lane 4). This pattern of cross-links demonstrates that catalytically important residues near the 5′ss and in J2/3 are very close in space and suggests a specific orientation for their interaction.
The third major target of cross-linking from the 5′-splice site are specific residues in D5, which are represented by cross-link group a (Fig. 4C,D). Specifically, s6dG1 forms a strong cross-link to position U814 (Fig. 4C), the nucleotide immediately preceding the universally conserved AGC “triad” of D5. In addition, the strong cross-links from s4U4 and s6dG6 map to A838 in the bulge of D5 (Fig. 4D). This catalytically important nucleotide is essential for splicing: modifications to its 2′OH (Abramovitz et al. 1996; Boudvillain and Pyle 1998), and to its nonbridging phosphate atoms (Chanfreau and Jacquier 1994; Boudvillain and Pyle 1998; Gordon and Piccirilli 2001) cause major defects in catalytic function. The observation of proximity between A838 and nucleotides C4 and G6 provides direct evidence for the location of this crucial nucleotide in relation to the tertiary structure of the group II intron.
Cross-links from photoactivated nucleotides in the bulge of Domain 5
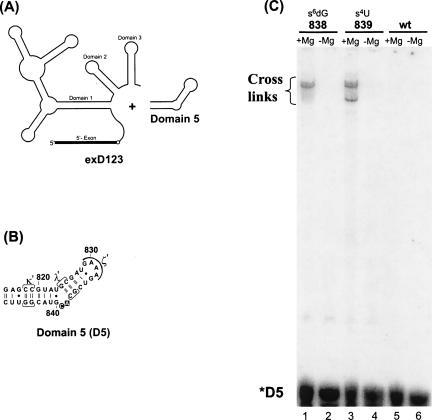
In order to further explore the proximity of catalytically essential elements and to corroborate the initial cross-linking data, we engineered an additional system whereby cross-linking analogs were placed in the opposite end of the molecule, within Domain 5 (D5). D5 molecules were synthesized with s4U and s6dG6 analogs in the bulge of D5, and then incubated with a transcript, exD123, which contains the 5′ exon as well as domains 1, 2, and 3. This trans system has been extensively utilized as a model system for quantitative analysis of the first step of group II intron splicing, whereby D5 catalyzes hydrolytic cleavage of exD123 with release of 5′ exon (Jarrell et al. 1988a; Franzen et al. 1993; Pyle and Green 1994).
Synthetic D5 molecules containing photoreactive nucleotides were all catalytically active, although s6dG modification at position 838 caused significant reduction in activity (15-fold reduction in kobs), which is consistent with previous work on the importance of the 2′OH at this position (Abramovitz et al. 1996). D5 molecules containing a s6dG analog at nucleotide 838 or a s4U analog at 839 formed strong cross-links with exD123 (Fig. 5). As in the previous case, cross-link formation depended on the presence of Mg++ and a photo-cross-linkable analog (Fig. 5C).
FIGURE 5.
Cross-linking from Domain 5. (A) The trans-hydrolysis system for monitoring the association of Domain 5 with an RNA containing the 5′ exon and domains 1, 2, and 3 (exD123) in a reaction that emulates the first step of splicing. (B) D5 oligonucleotides were synthesized with photo cross-linkable analogs s4U (circle) and s6dG (box) at the indicated positions. (C) 5′ 32P-labeled D5 oligonucleotides were preincubated with exD123 under trans-hydrolysis reaction conditions before exposure to 366 nm light. Mg++-dependent cross-links were observed for both D5 molecules containing cross-linkable analogs (lanes 1,3), but not for a wild type D5 (lane 5).
Reactivity of the cross-linked species was assessed by monitoring release of 5′ exon. Cross-links involving nucleotide 839 were fully competent for hydrolysis (data not shown). However, there was no detectable reactivity by cross-linked species involving nucleotide 838 (data not shown), which probably reflects the importance of the 2′OH moiety at that position and possible constraints on local conformation.
Mapping the spatial location of the D5 bulge
Cross-links from the bulge of D5 were mapped by RT primer extension, using primers complementary to exD123. The D5 variant containing s6dG838 formed a single, strong cross-link to position G5 of the intron (Fig. 6A), while D5 modified with s4U839 formed a strong cross-link to position G588 in the J2/3 linker (Fig. 6B). The proximity of A838 to G5 confirms that the D5 bulge is in close proximity to specific catalytic nucleotides near the 5′-splice site, which is consistent with the observation of cross-links from nucleotides 4 and 6 to A838 in the reverse experiment. The strong cross-link from 839 to G588 establishes another node in the tertiary arrangement of these regions: it demonstrates that the bulge of D5 is also in proximity of J2/3. The observation of a proximity constraint from D5 to J2/3 is consistent with previous work showing that J2/3 forms robust photo cross-links with D5 molecules that have been randomly substituted with s4U (Podar et al. 1998b). Together with the observation of cross-links from a photoactivated 5′-splice site to J2/3 and to the D5 bulge, these findings clearly indicate that three catalytically essential regions, i.e., the 5′ end of D1, J2/3, and D5 are in close proximity within the active architecture of the intron (within 9 Å). The specific pattern and differential arrangement of these cross-links allows us to place the bulge of D5 in its proper context within the active site.
FIGURE 6.
The D5 bulge cross-links to catalytically important residues. Mapping of D5 cross-links by reverse transcription primer extension. Cross-links are indicated by asterisks (*). (A) Nucleotide 838 in the D5 bulge cross-links to G5 near the 5′ splice site. (B) Nucleotide 839 in the D5 bulge cross-links to G588 in J2/3 linker region.
The results reported here are in accord with previous studies in which cross-linkers placed in D6 and the 3′ exon were used to visualize the organization of the intron active site (de Lencastre et al. 2005). These constraints, together with preliminary data on 5′-splice site and D5 cross-links (see Supplemental material; de Lencastre et al. 2005) enabled us to model a tertiary structure of the intron that is fully consistent with the analysis reported herein.
A functional role for the major groove atoms of G588 in J2/3
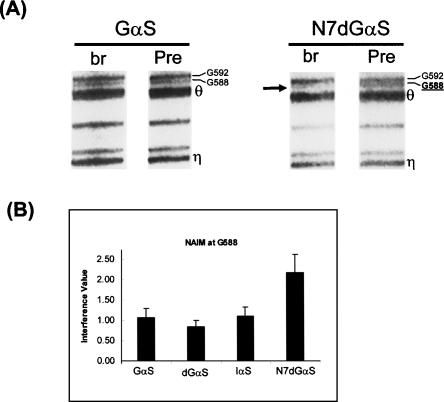
The cross-linking data are consistent with close proximity between nucleotide G588 in J2/3 and other active-site elements in the group II intron. Given the number of constraints that link catalytic elements with G588, we sought to further characterize the role of this nucleotide. In addition, it was important to determine whether the observed cross-links reflect functional interactions that are biologically relevant. To identify the functional groups that are important for catalytic function of G588, we employed NAIM (Conrad et al. 1995; Strobel and Shetty 1997). This technique allows one to probe the role of specific functional groups on all nucleotides of a given molecule without the laborious, and often impossible task of incorporating single-atom mutations. The approach was particularly useful for studying the J2/3 region, which is located in the middle of the intron and is therefore not highly amenable to semisynthetic procedures. We chose to probe major groove, minor groove, and sugar phosphate backbone functional groups for all guanosines in the ai5γ intron and focused specifically on the sensitivity of residues in J2/3. Transcription of exD123 was performed in the presence of αsGTP phosphorothioate analogs containing a variety of different single-atom modifications. The derivatives Guanosine-αs (Gαs), 2′deoxyG-αs (dGαs), inosine-αs (Iαs), and N7deazaG-αs (N7dGαs) were incorporated, and the well-characterized trans-branching system was used as the selection step for examining effects of these analogs (Chin and Pyle 1995; Boudvillain and Pyle 1998; Boudvillain et al. 2000). Analysis of the resultant products reveals that N7dGαs modification at position G588 has a very significant impact on trans-branching activity (Fig. 7), whereas other modifications, such as Gαs, dGαs, and Iαs, do not demonstrate noticeable effects (Fig. 7); Boudvillain and Pyle 1998). This result provides direct functional evidence that G588 plays a role during the first step of splicing, which is consistent with its demonstrable spatial proximity to elements near the 5′-splice site. Notably, it is the major groove N7 atom of G588 and not its minor groove or sugar moiety that is most important for catalysis. Taken together, our results demonstrate that the J2/3 region is both proximal to the 5′-splice site and catalytically relevant during the first step of splicing.
FIGURE 7.
Important functional groups on G588. Representative gel (A) and quantitation (B) of NAIM in the J2/3 region of the group II intron showing the specific N7dGαS interference effect at G588 (arrow). The labels “Pre” and “br” represent Precursor (input) and branched (selected) material, respectively. The labels Gαs, dGαs, Iαs, and N7dGαs denote exD123 transcripts containing phosphorothioate guanosine analogs G-αs, 2′deoxyG-αs, inosine-αs, and N7deazaG-αs, which probe the roles of the phosphate, the sugar, the minor, and major grooves of guanosine, respectively.
DISCUSSION
Identifying the spatial organization of group II intron active-site constituents
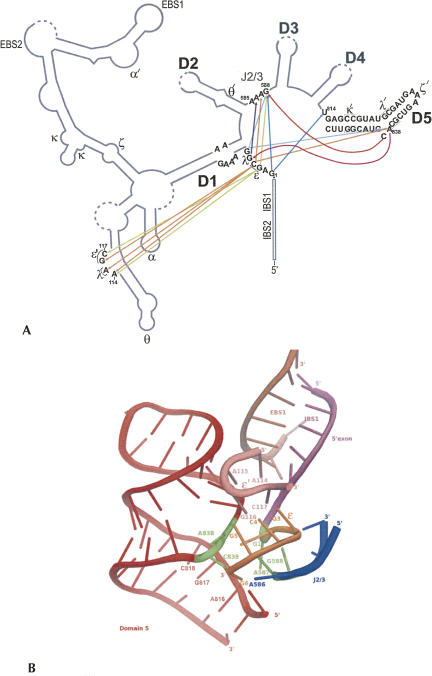
In this study, we utilized photo cross-linking to identify long-range tertiary interactions between catalytically essential and conserved regions of a group II intron (Fig. 8A). The short-range photo-cross-linkers s4U and s6dG were specifically incorporated near the 5′-splice site as well as in the internal bulge of Domain 5, and cross-linking formation was monitored in properly folded, catalytically active molecules. The cross-linking results indicate close spatial proximity of four major intronic regions (Fig. 8A,B), the first several nucleotides of the intron that are immediately downstream of the 5′-splice site, an internal bulge within the c1 stem of Domain 1, the conserved linker sequence between domains 2 and 3 (J2/3), and specific regions of the catalytically essential Domain 5. Importantly, most of these regions had been previously identified as critical for splicing and for catalytic activity of group II intron ribozymes (Jacquier and Michel 1990; Chanfreau and Jacquier 1994; Schmidt et al. 1996; Boudvillain and Pyle 1998; Boudvillain et al. 2000; Fedorova et al. 2003). The cross-linking results demonstrate that essential nucleotides in the 5′ end of D1, in D5, and in J2/3 are spatially colocalized in the vicinity of the 5′-splice site, suggesting that they comprise essential elements of the group II intron active site.
FIGURE 8.
Three-dimensional organization of the IIB intron. (A) Diagram summarizing the major cross-links observed in this study. Each cross-link donor (i.e., the incorporation site of a photo cross-linkable nucleotide) is color-coded and the arrowhead represents the site of attack. Straight lines indicate cross-links from the 5′ end of D1, while curved lines represent cross-links from the bulge of D5. (B) Active-site organization surrounding the 5′-splice site. Color scheme: 5′-exon (magenta); 5′ end of D1, including ε (orange), D1-c1 region, including ε′, EBS1 (brown), Domain 5 (red), and J2/3 (blue). The first nucleotide of the intron (G1), nucleotides of the D5 bulge, as well as A587 and G588 in J2/3 are highlighted in green. The data in this figure were adapted from the group IIB structural model (de Lencastre et al. 2005), with which all distance restraints reported here are fully consistent. Graphics were created using Pymol (DeLano 2002).
Characterizing the interactions and active-site role of the D5 bulge
The bulge substructure that separates the two helical stems of D5 is a critical component of the group II intron catalytic machinery and it bears structural and mechanistic similarities to an analogous bulge within the spliceosomal U2/U6 snRNA complex (Yean et al. 2000). The sequences of these bulges are semiconserved in both group II introns (5′-Ay) and in U2/U6 (5′-AU) and their deletion abolishes activity (Madhani et al. 1990; McPheeters 1996; Schmidt et al. 1996). Despite their importance, the active-site placement and catalytic role of the bulge nucleotides is not understood in either system. Here, we report the spatial location of the D5 bulge in relation to other catalytically important regions of the group II intron. Specifically, we find that the D5 bulge is proximal to two of the most conserved and catalytically essential regions of the group II intron: the nucleotides immediately downstream of the 5′-splice site and a highly conserved section of J2/3, indicating that these elements are proximal to the 5′-splice site during the first step of splicing (Fig. 8B).
The data indicate that one nucleotide in the D5 bulge (A838) is closely aligned with the conserved fifth nucleotide of the intron (G5), while the other bulge nucleotide (C839) is directed toward a different intronic element, J2/3. Consistent with this geometry, photoreactive groups placed at the 5′-splice site resulted in strong cross-links from the fourth and sixth nucleotides of the intron to A838, but not to C839. This pattern of cross-linking suggests that the bulge of D5 may have a bifurcated character, whereby A838 is oriented toward the 5′ terminus of the intron, while the major groove of C839 is oriented toward J2/3.
The cross-linked species examined in this study were shown to be catalytically active, indicating that they reflect a biologically relevant, active structure. In addition, the results are consistent with modeling studies in which each nucleotide of the D5 bulge was unstacked and pointing in a different direction (de Lencastre et al. 2005). All of these results suggest that the active conformation of the intron involves a D5 conformation in which A838 and C839 are extrahelical, allowing them to form contacts with the 5′ end of D1 and with nucleotides in J2/3, respectively. These results indicate that the bulge is an important element of the intron active site and they suggest possible parallels with analogous structures in the spliceosome.
The stem–loop structure within the spliceosomal U2/U6 complex and Domain 5 of the group II intron share many characteristics: they have similar secondary structures and similar levels of phylogenetic conservation and mutational sensitivity (for reviews, see Villa et al. 2002; Tycowski et al. 2006, and references within). Indeed, it has been shown that D5 from a group II intron can functionally replace this structure within the alternative, U12-dependent spliceosome (Shukla and Padgett 2002), which strongly suggests that the active sites of both machineries are structurally and mechanistically related. Consistent with this notion, phosphorothioate substitution at both C839 in the D5 bulge and the analogous U80 in spliceosomal U6 ablates catalysis in both systems (Fabrizio and Abelson 1992; Chanfreau and Jacquier 1994; Yu et al. 1995; Boudvillain and Pyle 1998; Yean et al. 2000; Gordon and Piccirilli 2001). Furthermore, the predicted secondary structures of D5 and U2/U6 indicate that both bulged nucleotides are spaced approximately the same distance (5 bp) from the highly conserved and catalytically essential triad of nucleotides (5′-AGC) that is located in the first stem. Finally, consistent with our cross-links between 5′ splice-site nucleotides and the D5 bulge, targeted hydroxyl radical experiments in activated spliceosomes have demonstrated proximity between the 10th nucleotide of a pre-mRNA and the bulge in the U6 ISL (Rhode et al. 2006).
Although these parallels are compelling, the active-site location of the bulge nucleotides has been unclear in both cases. In the spliceosome, a short-range cross-linker placed at U80 was found to cross-link with a nonconserved region of the pre-mRNA, and the relevance of this finding is unknown (Ryan et al. 2004). In contrast, we show a very strong, reproducible, and catalytically active cross-link from C839 to the highly conserved G588 in J2/3. These results suggest that, while spliceosomal U80 and group II intron C839 may both provide an essential role for catalysis through their phosphate backbones, the specific determinants for how they dock within the active site may differ. Alternatively, it may be that a spliceosomal counterpart for J2/3 exists, but has not yet been identified. In the spliceosomal case, this functionality might be provided by either RNA or protein components.
Nucleotides near the 5′-splice site interact with critical regions of the intron
The sequence downstream of the 5′-splice site in the group II introns is strongly conserved (5′-GNGYG) and bears a strong resemblance to the equivalent spliceosomal pre-mRNA sequence (5′-GURUG) (Cech 1993). In the spliceosome, this sequence was shown, first by cross-linking and later by genetics, to form a 2–3-bp intermolecular helix (5′-ss/U6) (Wassarman and Steitz 1992; Kandels-Lewis and Seraphin 1993; Lesser and Guthrie 1993), which might be analogous to the ε–ε′ interaction that has been described in the group II introns (Michel et al. 1989; Jacquier and Michel 1990). Our cross-linking results provide direct confirmation of this ε–ε′ interaction. Short-range cross-linkers placed at the third and fourth nucleotides of the intron form strong, antiparallel cross-links to G116 and C117 in the c1 stem of D1, consistent with their involvement in the ε–ε′ interaction.
The similarities of these sequences in the spliceosome and group II introns extend to their mutational sensitivity. Mutations to nucleotides near the 5′-splice site in the spliceosome strongly inhibit splicing (Woolford 1989). Similarly, both brute-force mutations as well as detailed functional characterization by NAIM show that the 5′ end of D1 in group II introns is highly sensitive to a wide variety of analog modifications in this region (Jacquier and Michel 1990; Chanfreau and Jacquier 1993; Peebles et al. 1993; Padgett et al. 1994; Podar et al. 1995a; Boudvillain and Pyle 1998; Boudvillain et al. 2000). The abundance and diversity of NAIM effects observed in the 5′ end of D1 suggests that this region coordinates a variety of long-range, multipartite interactions in the group II intron. Consistent with this, we find numerous long-range cross-links between the 5′ end of D1 and various regions of the intron. Combining the information obtained from cross-linking with previous studies allows us to propose a specific structural architecture for these catalytically essential regions and to discuss its functional implications.
One of the most critical nucleotides near the 5′-splice site is the highly conserved G5 adjacent to ε–ε′. Both mutational and NAIM studies indicate that minor groove, major groove, and sugar backbone functional groups of G5 are important for function (Boudvillain and Pyle 1998; Boudvillain et al. 2000). In the group II intron, NAIS experiments have shown that G5 forms an extended minor groove interaction, λ–λ′, which links the upper stem of D5 to D1, and the 5′-splice site. Indeed, when λ–λ′ was first identified, it was suggested that it might be part of a larger network that extended into the D5 bulge (Boudvillain et al. 2000), as shown here. The cross-linking results clearly demonstrate that, in the group II intron, Domain 5 is proximal to the 5′-splice site, in support of the previously identified λ–λ′ interaction. These results are further corroborated by the fact that s6dG placed at 838 in the bulge of D5 forms a strong, specific cross-link to G5, thereby confirming proximity between G5 and the bulge of Domain 5. In addition, in the reverse experiment, cross-linkers placed at nucleotides C4 and G6 near the 5′-splice site both cross-link to A838 in the D5 bulge. Thus, we provide evidence from two different experiments, with cross-linkers located at opposite ends of the molecule, that conclusively demonstrates proximity between the 5′-splice site and the D5 bulge in an active form of the molecule.
In spliceosomal introns, a similarly conserved G is located at the fifth position, raising the possibility of the existence of an analogous λ–λ′ interaction in the spliceosome. However, whereas G5 of group II introns forms a minor groove triple that bridges three distinct regions of the intron core, the spliceosomal G5 has only been shown, so far, to form a short duplex that is comparable to ε–ε′ in the group II intron. Such a short duplex is unlikely to be independently stable and is likely to depend on other interactions, perhaps necessitating protein stabilization or an undiscovered λ–λ′-like partner, as proposed previously (Boudvillain et al. 2000). It will therefore be very interesting to test whether the U6 analog of A838 is also proximal to G5, or to any other nucleotide in the spliceosomal sequence downstream of the 5′-splice site. Given that a test system already exists in the form of the U12 spliceosome (Shukla and Padgett 2002), a variety of different experimental approaches are possible for exploring this possibility.
G1: A novel role in the first step of splicing
Another important nucleotide at the 5′-splice site is G1, which is conserved and essential for both steps of splicing (Chanfreau and Jacquier 1993; Peebles et al. 1993). During the second step of splicing, G1 forms a noncanonical base pair with the penultimate nucleotide of the intron, an arrangement that resembles interactions between intronic boundaries in pre-mRNA splicing (Chanfreau and Jacquier 1993). However, any role for G1 in the first step of splicing has remained unclear. Its interaction with the penultimate nucleotide is observed only during the second step of splicing, which is implied by the fact that G1 mutations cause first-step defects that are unaffected by compensatory mutations in the penultimate nucleotide (Chanfreau and Jacquier 1993). Given these findings, it is likely that a hitherto unknown tertiary interaction involves G1 during the first step of splicing.
Here, we show that a s6dG analog at the first position of the intron cross-links efficiently with U814, which is the linker nucleotide immediately upstream of D5. Indeed, U814 is adjacent to the AGC “triad” of D5, which is among the most conserved and catalytically important regions of the intron (Chanfreau and Jacquier 1994; Boulanger et al. 1995; Peebles et al. 1995; Konforti et al. 1998a), and which resembles a similar triad within the spliceosomal U6 snRNA (Madhani and Guthrie 1992; Hilliker and Staley 2004). The conservation and mutational sensitivity of the AGC triad has long led to the expectation that it is a central component of the intron active site. However, to date, direct evidence for this hypothesis has been lacking. The cross-linking data reported herein represents important direct evidence of the proximity between the catalytic triad and the 5′-splice site.
Defining J2/3 as an active-site component
The purine-rich sequence (A/G)587 G588 A589 in J2/3 is almost invariant among group II introns, suggesting that it plays a critical role in function (Michel et al. 1989; Podar et al. 1998b). Indeed, deletions of J2/3 have been shown to considerably decrease the chemical-rate constants of both hydrolysis and branching reactions, indicating that it plays a fundamental role in catalysis (Fedorova et al. 2003). Nonetheless, the role of these nucleotides in catalysis and their location within the intron tertiary structure is not well understood.
Previous studies have shown that mutations to A587, G588, and A589 significantly impair the second step of splicing (Jacquier and Michel 1990; Mikheeva et al. 2000). On the basis of these studies, J2/3 has generally been assumed to play a role only in the second step. However, recent kinetic analyses have implicated an additional role in the first step of splicing. Mutations to G588 and A589 have been shown to significantly affect the first step of splicing, and the effects specifically impact catalysis and not binding (Ho Faix 1998). In support of a first-step role for J2/3, the structural data presented herein indicates that J2/3 is within the active site during the first step of catalysis. In ribozyme constructs that recapitulate only the first step of splicing, cross-links from nucleotides at the 5′ terminus of the intron and from the D5 bulge reproducibly target the conserved residues of J2/3. In NAIM studies that were designed to dissect the role of specific J2/3 functional groups during the first step of splicing, we observed that N7dG modification at G588, but not 2′deoxy, inosine, or Rp phosphorothioate, significantly impairs branching. This suggests that the major groove edge of G588 plays a key role within the intron active site, and that N7 has a specific functional role during the first step of splicing. Given that both J2/3 and C839 are strong and specific binding sites for divalent metal ions (Sigel et al. 2000), it will be interesting to determine whether their network of interactions is mediated by magnesium ions and/or whether they play a role in supporting metal ions that participate directly in catalysis.
Given the central role of J2/3 in both the first and second step of group II intron self-splicing, and a likely role for these nucleotides in chemical catalysis, it is important to consider the possible existence of a spliceosomal analog for this substructure. Specifically, it may be valuable to consider parallels between J2/3 and the conserved ACAGAGA sequence of U6 in the spliceosome. The trinucleotide A51G52A53 of U6 is highly conserved, like A587G588A589 of J2/3, and plays an important role in the second step of splicing (Jacquier and Michel 1990; Madhani and Guthrie 1994; Mikheeva et al. 2000). Structurally, it has been shown that A51 in U6 cross-links to the second nucleotide of the pre-mRNA (U2) near the 5′-splice site (Sontheimer and Steitz 1993), and genetic analysis indicates that this interaction is specifically important during the second step of splicing (Lesser and Guthrie 1993). Also during the second step, nucleotide G52 forms a tertiary interaction with A25 of U2, at the base of the U2/U6 hairpin stem (Madhani and Guthrie 1994). These results suggest a physical link between three spliceosomal regions: the 5′-splice site, the U2/U6 hairpin, and the A51G52A53 sequence. Similarly, we demonstrate in the group II intron that the 5′-splice site, the base of D5, and the AGA sequence in J2/3 are all in proximity. The similar structural disposition of these three regions in the spliceosome and in the group II intron is striking. Indeed, our structural model is quite similar to a model of the spliceosomal U2–U6 region comprising the pre-mRNA strand (with 5′-splice site), the ACAGAGA region, and the U6 ISL that is the putative analog of Domain 5 (Sashital et al. 2004). A detailed examination of a potential link between the spliceosomal AGA and J2/3 in group II introns may provide new insights into the relationship between group II introns and the modern spliceosome.
MATERIALS AND METHODS
Plasmids, templates, and RNA transcription
Plasmid pJDI3′-673 (Jarrell et al. 1988a), kindly provided by Dr. P.S. Perlman, encodes exD123 and was linearized by BamHI. Plasmid pT7-D56 (Chin and Pyle 1995), encoding D56, was linearized with DraI. Plasmid pexD135 (prepared by L.J. Su), encoding exD135, was linearized with HindIII. This construct is a variant of pQL71 (Swisher et al. 2001) containing a 293-nt-long 5′ exon, D1, D3, and D5 as well as shortened hairpin-loop replacements of D2 and D4. All plasmids were transcribed as previously described (Pyle and Green 1994; Chin and Pyle 1995). Site-specific modifications of the 5′ of D1 were introduced by treating exD135 transcripts with a 46-nt DNAzyme that was targeted to cleave between intron nucleotides G14 and U15, yielding RNA construct D135-14. This truncated molecule was then covalently joined, via template ligation (Moore and Sharp 1992), to a 40-nt synthetic RNA oligonucleotide (26.14) that corresponds to the first 26 nt of the 5′ exon and 14 nt of the intron (5′-UACUUACUACGUGGUGGGACAUUUUCGAGCGGUCUGAAAG), yielding a fully functional, semisynthetic 26D135 molecule.
Wild-type 26.14 and 34-mer D5 oligonucleotides were synthesized on a DNA/RNA synthesizer (ABI 392), deprotected, and purified according to published methods (Wincott et al. 1995). Oligonucleotides containing the photoactivated analogs s4U or s6dG (Glen Research) were obtained commercially (Dharmacon) (s4U) or synthesized in-house (s6dG). Oligonucleotides containing s4U and s6dG were deprotected according to the manufacturer's protocols, except that the base deprotection of s6dG-containing oligonucleotides involved the use of 0.05 M β-mercaptoethanol rather than 0.05 M NaSH. All steps of deprotection and oligonucleotide handling were performed in the dark.
RNA ligation, ribozyme preparation
Synthetic 26.14 oligonucleotides were joined to D135-14 RNA via template-directed ligation (Moore and Sharp 1992). Prior to ligation, D135-14 was phosphorylated using ATP and T4 polynucleotide kinase (New England Biolabs). The 5′-phosphorylated D135-14 was then ligated to 26.14 oligonucleotide using a 32-nt DNA splint (5′-GTAAATATTATTTATGATAACTTTCAGACCGC) and T4 DNA ligase (New England Biolabs), resulting in a 26D135 ribozyme. Ligations were performed for 4 h at 30°C in a 150-μL reaction volume using 20,000 U of T4 DNA ligase, 400 U of RNAsin (Roche), and 1× T4 DNA ligase buffer (New England Biolabs). Ligated RNAs were then isolated by 5% denaturing PAGE. Ligations were efficient (∼40%), and the final yield of ligated construct relative to starting transcript averaged 10%–20% after all purification steps.
Kinetics
To evaluate whether constructs retained the ability to undergo the first step of splicing, 5′ end-labeled exD135 or 26D135 molecules (1–10 nM) were added to a buffer containing 0.5 M KCl, 80 mM MOPS (pH 7.5) in 10 μL reaction volume. This solution was heated at 95°C for 1 min and then allowed to fold for 5 min at 42°C. Reaction was initiated by the addition of 0.1 M MgCl2 (final concentration). At selected time intervals, aliquots were withdrawn and combined with 2 vol of quench buffer (95% formamide, 5 mM EDTA (pH 6.0), 0.1% of xylene cyanol and bromophenol blue dyes) before placing on ice. Samples were loaded on 5% PAGE and analyzed by PhosphorImager (Amersham Biosciences). The data were fit to a simple exponential equation: frac (P) = 1 − e−kt, where P represents product, t is time, and k is the observed rate constant (min−1).
Cross-linking
To monitor the evolution of cross-links from the 5′ terminus of D1, 75 pmols of 5′ end-labeled 26D135 were dissolved in a buffer of 80 mM MOPS (pH 7.5) and 0.5 M KCl. This solution was heated at 95°C for 1 min, cooled at room temperature for 1 min, and then combined with 0.1 M MgCl2 for a final reaction volume of 10 μL. After incubating the 26D135 molecules for 5 min at 42°C, the samples were then transferred to a covered crystallography tray (Hampton Research) and placed under a UV-light source (Hand-held UV illuminator, model UVGL-58) set at λ = 366 nm for 30 min. Cross-linked material was then separated from precursor molecules and purified by denaturing PAGE.
To monitor the evolution of cross-links from D5, conditions were similar to those described above. 5′ end-labeled D5 (75 pmols) and exD123 (5 μM) were denatured separately at 95°C for 1 min and cooled at room temperature for 1 min in reaction buffer containing in 0.5 M KCl and 80 mM MOPS (pH 7.5). They were then combined and 0.1 M MgCl2 was added. The complex was allowed to incubate for 15 min at 45°C before the samples were irradiated and isolated as described above. All steps involving cross-linkable material were performed in the dark.
Evaluating the reactivity of cross-linked ribozyme molecules
The radiolabeled, cross-linked RNA products were eluted from gel slices, ethanol precipitated, and resuspended in reaction buffer (0.5 M KCl, 80 mM MOPS at pH 7.5). The samples were then denatured at 95°C for 1 min, cooled at room temperature for 1 min, and then combined with 0.1 M MgCl2 for a final reaction volume of 10 μL. Samples were then incubated at 42°C and aliquots were withdrawn at selected time points and combined with 2 vol of quench buffer (95% formamide, 5 mM EDTA at pH 6.0, 0.1% of xylene cyanol and bromophenol blue dyes). Samples were then subjected to electrophoresis on a 5%:20% (top:bottom) stacked denaturing polyacrylamide gel in order to separate reaction products and to observe release of the 26-nt 5′ exon. As a control, 5′ end-labeled 26D135 (wt) was simultaneously tested.
RT mapping of cross-links
RT-AMV (Roche) was used to map the location of cross-links, following standard RT mapping procedures (Stern et al. 1988; Konforti et al. 1998b; Juzumiene et al. 2001). Several DNA primers complementary to different regions of the RNA were used to scan the entire intron. In this procedure, cross-linked RNA species were resuspended in 20 μL of storage buffer (ME, pH 6.0) and 4 μL was aliquoted to each of five different RT reactions. A total of 50 pmol of a 5′ end-labeled DNA primer was added to each RT reaction, and the mixture was denatured at 90°C for 1 min. The sample was then allowed to slow cool to 55°C and the RT reaction was initiated by addition of 1X AMV-RT Reaction Mix (Roche), 0.7 mM dNTP mix, 10 U of RNasin (Roche), and 6 U of RT-AMV (Roche) in a final reaction volume of 10 μL. For dideoxy sequencing, separate RT reactions were performed on unmodified 26D135 RNA in the presence of 0.05 mM of each ddNTP. All RT reactions were allowed to proceed for 15 min at 55°C and were stopped by the addition of an equal volume of urea/sucrose loading buffer. Prior to loading on an 8% sequencing gel, samples were heated to 90°C for 1 min. All gels were dried and analyzed on a PhosphorImager (Amersham Biosciences). The location of a cross-link was determined by comparison to a dideoxy sequencing pattern: a cross-link will cause the RT to stop one base prior to the corresponding position on a dideoxy sequencing ladder (Sanger et al. 1977). Nonirradiated precursor (pre) RNA was used as a control for the presence of natural RT stops.
Nucleotide analog interference mapping
All GαS, dGαS, IαS, and N7dGαS modifications were randomly incorporated into exD123 at a 5% incorporation level by transcription (Ryder and Strobel 1999). The pool of modified exD123 molecules was subjected to a trans-branching reaction with D56 (Chin and Pyle 1995), which represents the selection step for the NAIM analysis, as described (Boudvillain and Pyle 1998). Specifically, NαS-modified exD123 (1.5 μM) was combined with 5′ end-labeled D56 (1–10 nM), in reaction buffer (0.5 M NH4Cl, 40 mM MOPS at pH 6.0) and heated for 1 min at 90°C. After cooling at room temperature for 1 min, 0.1 M MgCl2 was added for a final reaction volume of 12 μL. The samples were then incubated at 42°C, and reaction time was adjusted so that the final reaction extent was ∼20% (reacted/total). The reaction was stopped by adding an equal volume of formamide loading buffer, and the samples were loaded on a 5% denaturing polyacrylamide gel. Branched species were isolated from the gel, eluted, and ethanol precipitated before sequencing. For sequencing, precursor and branched species were incubated with 10 mM I2 in a 6 μL reaction volume for 3 min at 37°C. An equal volume of urea/sucrose loading buffer was added and the samples were sequenced by 5% PAGE. All gels were dried and analyzed on a PhosphorImager (Amersham Biosciences). Interference quantitation is performed as previously described (Ortoleva-Donnelly et al. 1998), and the interference values represent the average of three independent experiments (error did not exceed 20%). The interference values were normalized for loading differences and the reported interference values represent the average of two independent experiments.
ACKNOWLEDGMENTS
We thank Olga Fedorova for RNA synthesis and help with photo cross-linking, as well as Stephanie Hamill and Philip S. Perlman for helpful discussions. This work was generously supported by a grant from the National Institutes of Health (GM50313 to A.M.P.) and by fellowships from Praxis XXI and Gulbenkian 43058 (to A.d.L.). A.M.P. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.774008.
REFERENCES
- Abramovitz, D.L., Friedman, R.A., Pyle, A.M. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science. 1996;271:1410–1413. doi: 10.1126/science.271.5254.1410. [DOI] [PubMed] [Google Scholar]
- Bonen, L., Vogel, J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- Boudvillain, M., Pyle, A.M. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: A NAIM analysis. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain, M., de Lencastre, A., Pyle, A.M. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- Boulanger, S.C., Belcher, S.M., Schmidt, U., Dib-Hajj, S.D., Schmidt, T., Perlman, P.S. Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron. Mol. Cell. Biol. 1995;15:4479–4488. doi: 10.1128/mcb.15.8.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech, T.R. Structure and mechanism of the large catalytic RNAs: Group I and Group II introns and ribonuclease P. In: Gesteland R.F., Atkins J., editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 239–269. [Google Scholar]
- Chanfreau, G., Jacquier, A. Interaction of intronic boundaries is required for the second splicing step efficiency of a group II intron. EMBO J. 1993;12:5173–5180. doi: 10.1002/j.1460-2075.1993.tb06212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Jacquier, A. Catalytic site components common to both splicing steps of a group II intron. Science. 1994;266:1383–1387. doi: 10.1126/science.7973729. [DOI] [PubMed] [Google Scholar]
- Chin, K., Pyle, A.M. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- Conrad, F., Hanne, A., Gaur, R.K., Krupp, G. Enzymatic synthesis of 2′-modified nucleic acids: Identification of important phosphate and ribose moieties in RNase P substrates. Nucleic Acids Res. 1995;23:1845–1853. doi: 10.1093/nar/23.11.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M., Michel, F., Westhof, E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000;19:5007–5018. doi: 10.1093/emboj/19.18.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, D.L., Michels W.J., Jr, Pyle, A.M. Two competing pathways for self-splicing by group II introns: A quantitative analysis of in vitro reaction rates and products. J. Mol. Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- de Lencastre, A., Hamill, S., Pyle, A.M. A single active-site region for a group II intron. Nat. Struct. Mol. Biol. 2005;12:626–627. doi: 10.1038/nsmb957. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. The Pymol molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Fabrizio, P., Abelson, J. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 1992;20:3659–3664. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova, O., Mitros, T., Pyle, A.M. Domains 2 and 3 interact to form critical elements of the group II intron active site. J. Mol. Biol. 2003;330:197–209. doi: 10.1016/s0022-2836(03)00594-1. [DOI] [PubMed] [Google Scholar]
- Franzen, J.S., Zhang, M., Peebles, C.L. Kinetic analysis of the 5′ splice junction hydrolysis of a group II intron promoted by domain 5. Nucleic Acids Res. 1993;21:627–634. doi: 10.1093/nar/21.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, P.M., Piccirilli, J.A. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat. Struct. Biol. 2001;8:893–898. doi: 10.1038/nsb1001-893. [DOI] [PubMed] [Google Scholar]
- Gumbs, O.H., Padgett, R.A., Dayie, K.T. Fluorescence and solution NMR study of the active site of a 160-kDa group II intron ribozyme. RNA. 2006;12:1693–1707. doi: 10.1261/rna.137006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill, S., Pyle, A.M. The receptor for branch-site docking within a group II intron active site. Mol. Cell. 2006;23:831–840. doi: 10.1016/j.molcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Hilliker, A.K., Staley, J.P. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Faix, P. Conserved nucleotides in the joining segment between Domains 2 and 3 are important for Group II splicing. University of Pittsburgh; Pittsburgh, PA: 1998. [Google Scholar]
- Jacquier, A., Michel, F. Base-pairing interactions involving the 5′- and 3′-terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- Jarrell, K.A., Dietrich, R.C., Perlman, P.S. Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell. Biol. 1988a;8:2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell, K.A., Peebles, C.L., Dietrich, R.C., Romiti, S.L., Perlman, P.S. Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem. 1988b;263:3432–3439. [PubMed] [Google Scholar]
- Juzumiene, D., Shapkina, T., Kirillov, S., Wollenzien, P. Short-range RNA–RNA cross-linking methods to determine rRNA structure and interactions. Methods. 2001;25:333–343. doi: 10.1006/meth.2001.1245. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis, S., Seraphin, B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Koch, J.L., Boulanger, S.C., Dib-Hajj, S.D., Hebbar, S.K., Perlman, P.S. Group II introns deleted for multiple substructures retain self-splicing activity. Mol. Cell. Biol. 1992;12:1950–1958. doi: 10.1128/mcb.12.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti, B.B., Abramovitz, D.L., Duarte, C.M., Karpeisky, A., Beigelman, L., Pyle, A.M. Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell. 1998a;1:433–441. doi: 10.1016/s1097-2765(00)80043-x. [DOI] [PubMed] [Google Scholar]
- Konforti, B.B., Liu, Q., Pyle, A.M. A map of the binding site for catalytic domain 5 in the core of a group II intron ribozyme. EMBO J. 1998b;17:7105–7117. doi: 10.1093/emboj/17.23.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, K., Schmidt, U. Group II introns: Structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- Lesser, C.F., Guthrie, C. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Guthrie, C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Guthrie, C. Randomization-selection analysis of snRNAs in vivo: Evidence for a tertiary interaction in the spliceosome. Genes & Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., Bordonne, R., Guthrie, C. Multiple roles for U6 snRNA in the splicing pathway. Genes & Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- McPheeters, D.S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., Ozeki, H. Comparative and functional anatomy of group II catalytic introns–A review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Michels W.J., Jr, Pyle, A.M. Conversion of a group II intron into a new multiple-turnover ribozyme that selectively cleaves oligonucleotides: Elucidation of reaction mechanism and structure/function relationships. Biochemistry. 1995;34:2965–2977. doi: 10.1021/bi00009a028. [DOI] [PubMed] [Google Scholar]
- Mikheeva, S., Murray, H.L., Zhou, H., Turczyk, B.M., Jarrell, K.A. Deletion of a conserved dinucleotide inhibits the second step of group II intron splicing. RNA. 2000;6:1509–1515. doi: 10.1017/s1355838200000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Sharp, P.A. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Noah, J., Lambowitz, A. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003;42:12466–12480. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- Ortoleva-Donnelly, L., Szewczak, A.A., Gutell, R.R., Strobel, S.A. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett, R.A., Podar, M., Boulanger, S.C., Perlman, P.S. The stereochemical course of group II intron self-splicing. Science. 1994;266:1685–1688. doi: 10.1126/science.7527587. [DOI] [PubMed] [Google Scholar]
- Peebles, C.L., Benatan, E.J., Jarrell, K.A., Perlman, P.S. Group II intron self-splicing: Development of alternative reaction conditions and identification of a predicted intermediate. Cold Spring Harb. Symp. Quant. Biol. 1987;52:223–232. doi: 10.1101/sqb.1987.052.01.027. [DOI] [PubMed] [Google Scholar]
- Peebles, C.L., Belcher, S.M., Zhang, M., Dietrich, R.C., Perlman, P.S. Mutation of the conserved first nucleotide of a group II intron from yeast mitochondrial DNA reduces the rate but allows accurate splicing. J. Biol. Chem. 1993;268:11929–11938. [PubMed] [Google Scholar]
- Peebles, C.L., Zhang, M., Perlman, P.S., Franzen, J.S. Catalytically critical nucleotides in domain 5 of a group II intron. Proc. Natl. Acad. Sci. 1995;92:4422–4426. doi: 10.1073/pnas.92.10.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar, M., Dib-Hajj, S., Perlman, P.S. A UV-induced, Mg2+-dependent cross-link traps an active form of domain 3 of a self-splicing group II intron. RNA. 1995a;1:828–840. [PMC free article] [PubMed] [Google Scholar]
- Podar, M., Perlman, P.S., Padgett, R.A. Stereochemical selectivity of group II intron splicing, reverse splicing, and hydrolysis reactions. Mol. Cell. Biol. 1995b;15:4466–4478. doi: 10.1128/mcb.15.8.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar, M., Perlman, P.S., Padgett, R.A. The two steps of group II intron self-splicing are mechanistically distinguishable. RNA. 1998a;4:890–900. doi: 10.1017/s1355838298971643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar, M., Zhuo, J., Zhang, M., Franzen, J.S., Perlman, P.S., Peebles, C.L. Domain 5 binds near a highly conserved dinucleotide in the joiner linking domains 2 and 3 of a group II intron. RNA. 1998b;4:151–166. [PMC free article] [PubMed] [Google Scholar]
- Pyle, A.M., Green, J.B. Building a kinetic framework for group II intron ribozyme activity: Quantitation of interdomain binding and reaction rate. Biochemistry. 1994;33:2716–2725. doi: 10.1021/bi00175a047. [DOI] [PubMed] [Google Scholar]
- Pyle, A.M., Lambowitz, A.M. Group II introns: Ribozymes that splice RNA and invade DNA. In: Gesteland R.F., Cech T.R., Atkins J., editors. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 469–505. [Google Scholar]
- Qin, P.Z., Pyle, A.M. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- Rhode, B.M., Hartmuth, K., Westhof, E., Luhrmann, R. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, D.E., Kim, C.H., Murray, J.B., Adams, C.J., Stockley, P.G., Abelson, J. New tertiary constraints between the RNA components of active yeast spliceosomes: A photo-cross-linking study. RNA. 2004;10:1251–1265. doi: 10.1261/rna.7060404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, S.P., Strobel, S.A. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital, D.G., Cornilescu, G., McManus, C.J., Brow, D.A., Butcher, S.E. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Schmidt, U., Podar, M., Stahl, U., Perlman, P.S. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: Phenotypes and suppressor mutations. RNA. 1996;2:1161–1172. [PMC free article] [PubMed] [Google Scholar]
- Seetharaman, M., Eldho, N.V., Padgett, R.A., Dayie, K.T. Structure of a self-splicing group II intron catalytic effector domain 5: Parallels with spliceosomal U6 RNA. RNA. 2006;12:235–247. doi: 10.1261/rna.2237806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, G.C., Padgett, R.A. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol. Cell. 2002;9:1145–1150. doi: 10.1016/s1097-2765(02)00505-1. [DOI] [PubMed] [Google Scholar]
- Sigel, R.K., Vaidya, A., Pyle, A.M. Metal ion binding sites in a group II intron core. Nat. Struct. Biol. 2000;7:1111–1116. doi: 10.1038/81958. [DOI] [PubMed] [Google Scholar]
- Sigel, R.K., Sashital, D.G., Abramovitz, D.L., Palmer, A.G., Butcher, S.E., Pyle, A.M. Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif. Nat. Struct. Mol. Biol. 2004;11:187–192. doi: 10.1038/nsmb717. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Steitz, J.A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Stern, S., Moazed, D., Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Strobel, S.A., Shetty, K. Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. Proc. Natl. Acad. Sci. 1997;94:2903–2908. doi: 10.1073/pnas.94.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher, J., Duarte, C.M., Su, L.J., Pyle, A.M. Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J. 2001;20:2051–2061. doi: 10.1093/emboj/20.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher, J.F., Su, L.J., Brenowitz, M., Anderson, V.E., Pyle, A.M. Productive folding to the native state by a group II intron ribozyme. J. Mol. Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- Toor, N., Hausner, G., Zimmerly, S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, N. Bacteria and Archaea Group II introns: Additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., Kolev, N.G., Conrad, N.K., Fok, V., Steitz, J.A. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland R.F., et al., editors. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 327–368. [Google Scholar]
- Villa, T., Pleiss, J.A., Guthrie, C. Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell. 2002;109:149–152. doi: 10.1016/s0092-8674(02)00726-2. [DOI] [PubMed] [Google Scholar]
- Wassarman, D.A., Steitz, J.A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Wincott, F., DiRenzo, A., Shaffer, C., Grimm, S., Tracz, D., Workman, C., Sweedler, D., Gonzalez, C., Scaringe, S., Usman, N. Synthesis, deprotection, analysis, and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J.L., Jr Nuclear pre-mRNA splicing in yeast. Yeast. 1989;5:439–457. doi: 10.1002/yea.320050604. [DOI] [PubMed] [Google Scholar]
- Yean, S.L., Wuenschell, G., Termini, J., Lin, R.J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Maroney, P.A., Darzynkiwicz, E., Nilsen, T.W. U6 snRNA function in nuclear pre-mRNA splicing: A phosphorothioate interference analysis of the U6 phosphate backbone. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Doudna, J.A. Structural insights into group II intron catalysis and branch-site selection. Science. 2002;295:2084–2088. doi: 10.1126/science.1069268. [DOI] [PubMed] [Google Scholar]