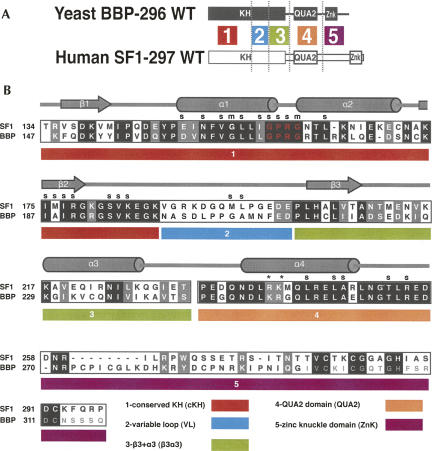
FIGURE 2.
A map of the five regions used to make the BBP and SF1 chimeras. (A) The domain structure of BBP-296 (black) and SF1-297 (white), and placement of the five different regions. (B) An alignment of BBP-296 and SF1-297 and detailed map of the five regions underneath. A key of the five regions and their names are listed at the bottom of the alignment. In the alignment, black boxes show identical amino acids between BBP and SF1, and gray boxes represent similar amino acids between the two proteins. The GXXG motif is shown in red letters. Secondary structure and contacts to the RNA are marked (s for side chain and m for main chain RNA contacts, cylinders for α-helices, and arrows for β-strands) based on the human SF1 NMR structure (Liu et al. 2001). An asterisk (*) above the alignment marks the important arginine (R) and lysine (K) discussed in Figure 6. The chimeras were named according to the region of the protein that was replaced with the yeast sequence and a y placed before the region name to represent yeast. For example, in the construct SF1-ycKH, the yeast-conserved KH (cKH) region was exchanged for the homologous human cKH region. The second zinc knuckle of BBP was not used in the protein constructs for binding studies, and those amino acids are shown in a smaller light gray in the alignment.