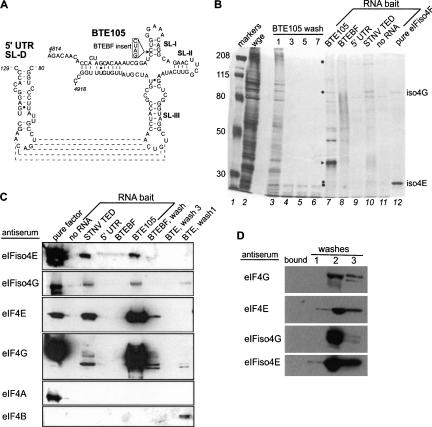
FIGURE 1.
Identification of BTE-interacting proteins from wheat germ extract. (A) Secondary structure of BTE105 RNA (right) and the stem–loop in the 5′UTR (SL-D) to which it base pairs (indicated by dashed lines) (Guo et al. 2001). In mutant BTEBF (inset), the BamHI4837 site (nucleotide 4837) is disrupted by insertion of a GAUC duplication that abolishes BTE function. (B) BTE-interacting proteins (BTEIPs) from wheat germ extract (wge) identified using biotin-labeled RNAs as bait and magnetic streptavidin beads to pull down the BTEIPs. Proteins were separated by 5% PAGE and silver stained. (Lane 1) Molecular weight markers of the indicated kDa at left; (lane 2) total wge protein; (lanes 3–6) unbound proteins obtained after indicated number of low-salt washes of bead-bound BTE RNA. RNA bait (lanes 7–11) show proteins that remained bound to the indicated RNA after washes. STNV TED consists of nucleotides 621–741 of STNV RNA. (Lane 12) Recombinant eIFiso4E and eIFiso4G. Dots to the left of lane 7 indicate BTE105-interacting proteins that comigrate with eIFiso4E and eIFiso4G. Expected Mobilities of eIF4G (> 200 kDa) and eIF4E (26 kDa) are indicated by squares. Arrowhead indicates BTE RNA. (C) Western blots using antibodies to known initiation factors on proteins pulled down by the indicated RNAs. Proteins were separated by SDS PAGE prior to blotting on PVDF membrane. Each panel represents a different gel and blot. Washes indicate proteins not bound to the indicated RNA. No pure eIF4B was used as positive control (first lane), but efficacy of antisera was evident by detection of eIF4B in the low-salt wash. Cleavage products of the labile eIF4G are visible. (D) Western blots against wheat germ extract proteins eluted from biotinylated, nonviral (166 nt vector-derived) RNA complexes. (bound) Proteins interacting with vector sequence (none detected in these Western blots). Unbound proteins obtained in washes (lanes 1–3) as indicated.