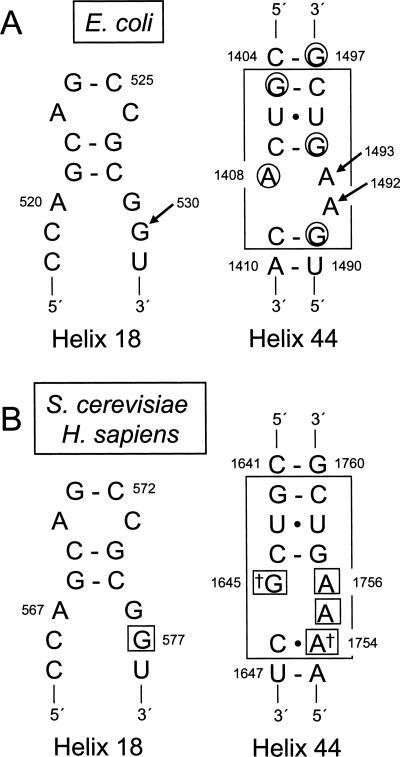
FIGURE 1.
Decoding site residues from E. coli 16S rRNA and S. cerevisiae and H. sapiens 18S rRNAs. (A) Secondary structures of the portions of helices 18 and 44 of E. coli 16S rRNA that contribute to the decoding site. The portion of helix 18 containing residue G530 is shown on the left. Residues of the core decoding stem in helix 44 are boxed on the right (Pfister et al. 2003a). Bases protected from DMS modification by paromomycin are circled (Yoshizawa et al. 1998). Arrows indicate the decoding site residues G530, A1492, and A1493. (B) Secondary structures of corresponding portions of helices 18 and 44 in S. cerevisiae and H. sapiens 18S rRNAs. Residues corresponding to the core decoding stem in E. coli helix 44 are enclosed by the large box. Daggers mark the two bases of the decoding site not conserved between prokaryotes and eukaryotes. Small boxes indicate the residues mutated in this study.