Figure 1.
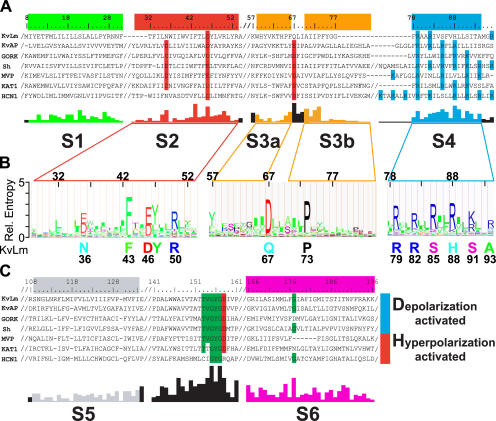
A prokaryotic Kv channel with essential sequence conservation of the voltage sensor. (A and C) Representative Clustal X alignments of the voltage sensor module (A) and pore module (C) sequence of KvLm with sequences of six known Kv channels from bacteria (KvAP and MVP), plant (GORK and KAT1), and other eukaryotes (Sh and HCN1) (A and C). Conserved residues, functional in voltage sensing or K+ selectivity, are shaded red (acidic), blue (basic), and green (uncharged). Residue numbers are for KvLm; “/” indicates sequence breaks. Abbreviated names, accession nos., and genome source are as follows: KvLm, gi 16411529, Listeria monocytogenes; MVP, gi 15668309, Methanococcus jannaschii; KAT1, gi 10177705, Arabidopsis thaliana; HCN1, gi 29840778, Mus musculus; KvAP, gi 14601099, Aeropyrum pernix; GORK, gi 30693099, Arabidopsis thaliana; Sh, gi 13432103, Drosophila melanogaster. The conservation score below the alignment was calculated for the master alignment encompassing 19 known Kv channels and 54 Kv channel candidates from prokaryotic genomes (Fig. S1). Color bars (green for S1, red for S2, yellow for S3a and S3b, blue for S4, gray for S5, and magenta for S6) above sequence alignment and colored conservation score plot under alignment delimit transmembrane α-helices as inferred from the structure of KvAP (Jiang et al., 2003a; Cuello et al., 2004). (B) Hidden Markov model (HMM) profile for S2, S3, and S4 calculated from master alignment. The height of the residues is proportional to conservation. Below profile, at sequence positions found to be highly conserved (relative entropy > 2), the corresponding residue in KvLm is shown.