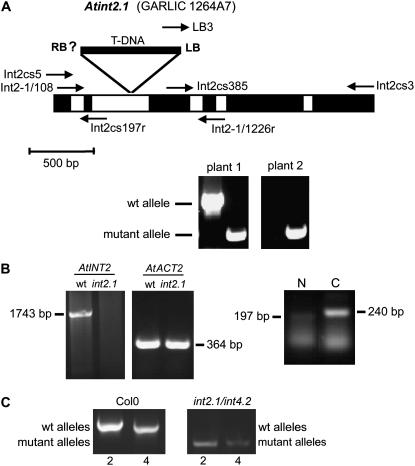
Figure 8.
Characterization of GARLIC line 1264A7 (=Atint2.1). A, Schematic drawing of the AtINT2 genomic sequence from start to stop. Exons are shown in black, the five introns in white. The position of the T-DNA insertion and of the characterized left border (LB) are indicated. Arrows show the positions and directions of primers used for PCR reactions in A and B. Example PCRs for two progeny plants of GARLIC_1264A7 seeds identifying wild-type (primers: Int2-1/108 and Int2-1/1226r) and mutant (primers: LB3 and Int2-1/1226r) alleles are presented. Plant 1 was characterized as heterozygous, plant 2 as homozygous. B, RT-PCR-derived bands showing the complete AtINT2 ORF (primers: Int2cs5 and Int2cs3) and the partial AtACT2 ORF (primers: ACT2-846f and ACT2-1295r) from RNA isolated from wild-type or Atint2.1 plants. While an AtINT2 product was obtained only from wild-type plants, the ACT2 fragment was amplified both from wild-type and mutant plants. cDNA fragments were amplified from truncated mRNAs upstream and downstream of the insertion site (N: primers Int2cs5 and Int2cs197r; C: primers Int2cs385 and Int2-1/1226r). C, Characterization of an Atint2.1/Atint4.2 double mutant. Only in Col-0 but not in the double mutant the wild-type alleles of AtINT2 and AtINT4 were identified (primers: Int2-1/108 and Int2-1/1226r for AtINT2; int4-802 and int4-1826r for AtINT4; Schneider et al. [2006]). In contrast, mutant alleles were amplified only in the double mutant (primers: LB3 and Int2-1/1226r for Atint2.1; int4-802 and LBb1 for Atint4.2; Schneider et al. [2006]).