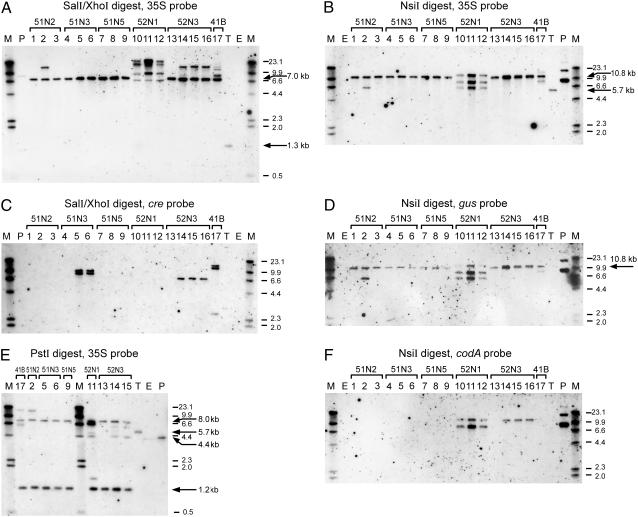
Figure 3.
Southern-blot analyses of putative RMCE lines. Chromosomal DNA of target plant (T), untransformed control ecotype C24 (E), and plants of putative RMCE lines derived from experiments 4 and 5 was either digested with NsiI (B, D, and F) or PstI (E) or double digested with SalI and XhoI (A and C), and hybridized with 35S (A, B, and E), cre (C), gus (D), or codA (F) probes, respectively. Codes above the horizontal brackets indicate the name of the parent plant line of which the individuals in the lanes are derived. Numbering of the individuals is consistently used for all blots. The name of the line indicates the following. First identifier, Experiment number; second identifier, 1 for exchange T-DNA 3327 or 2 for T-DNA 3732; third identifier, N for T-DNA containing pnos-cre or B for pBigmac-cre. When more than one callus was selected, the last identifier indicates the callus number of which the plants originated. Individual lanes underneath a bracket show different kanamycin-resistant progeny plants from a single regenerated R1 shoot. Lane P contains digested plasmid DNA of exchange construct pSDM3327 mixed with digested C24 chromosomal DNA; M, DIG-labeled molecular weight marker. Arrows indicate the expected RMCE fragment 7.0 kb and the target fragment 1.3 kb (A); the expected RMCE fragment 10.8 kb and the target fragment 5.7 kb (B); the expected 10.8-kb RMCE fragment (D); and CE fragments of 1.2 kb and 8.0 kb, the target fragment 5.7 kb, and the 4.4-kb fragment specific for exchange T-DNA, not recombined at the lox5171 site (E).