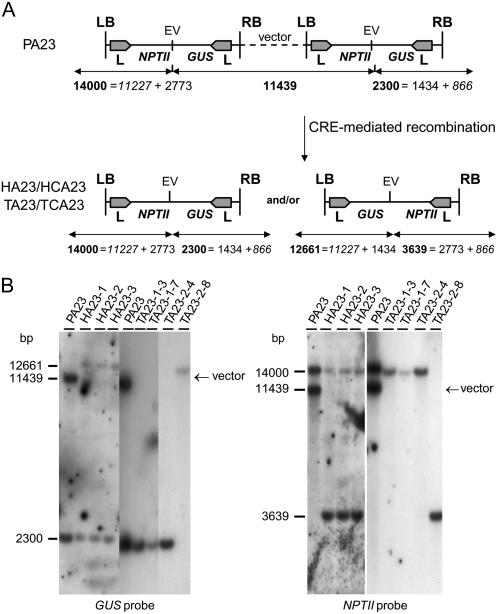
Figure 2.
DNA gel-blot analysis on EcoRV-digested DNA of PA23, HA23, and TA23. A, Schematic representation of the complex locus PA23 and the resolved/inverted T-DNA in HA23/HCA23 and TA23/TCA23 seedlings. The PA23 transformant carried two T-DNAs in direct orientation with vector backbone sequences in between, resulting in two fragments after hybridization with the GUS (11,439 bp and 2,300 bp) and NPTII probes (11,439 bp and 14,000 bp). For the two T-DNA/plant junctions, the T-DNA fragment was known (2,773 bp and 1,434 bp for the LB and RB regions, respectively; Fig. 1), and the plant fragment, in italics, could be calculated (11,227 bp and 866 bp for the LB and RB plant regions, respectively). After resolution of the complex T-DNA locus, two different T-DNA configurations could be found: the original T-DNA configuration with the same T-DNA/plant junction fragments as in the parental line and the inverted T-DNA configuration. In this latter configuration, the length of the fragments could be predicted again because both T-DNA and plant fragments of the junctions were known: for the NPTII and GUS probes, a fragment of 3,639 bp (2,773 bp of the NPTII T-DNA fragment and 866 bp for the RB/plant junction) and a fragment of 12,661 bp (11,227-bp LB/plant junction + 1,434-bp T-DNA fragment), respectively. B, DNA gel-blot analysis on DNA of parental (P), hybrid (H), and progeny (T) plants of A23 with the GUS (left) and NPTII probes (right). The hybrids were obtained after crossing the parental plants with the CRE13 line. On top of each lane, the name is given of the transformant from which DNA was prepared. Abbreviations: EV, EcoRV; L, loxP sequence recognition site of the Cre/loxP recombination system; vector, read through at the LB results in a tandem repeat of two Ksb T-DNAs with the entire vector backbone sequence in between.