Figure 1.
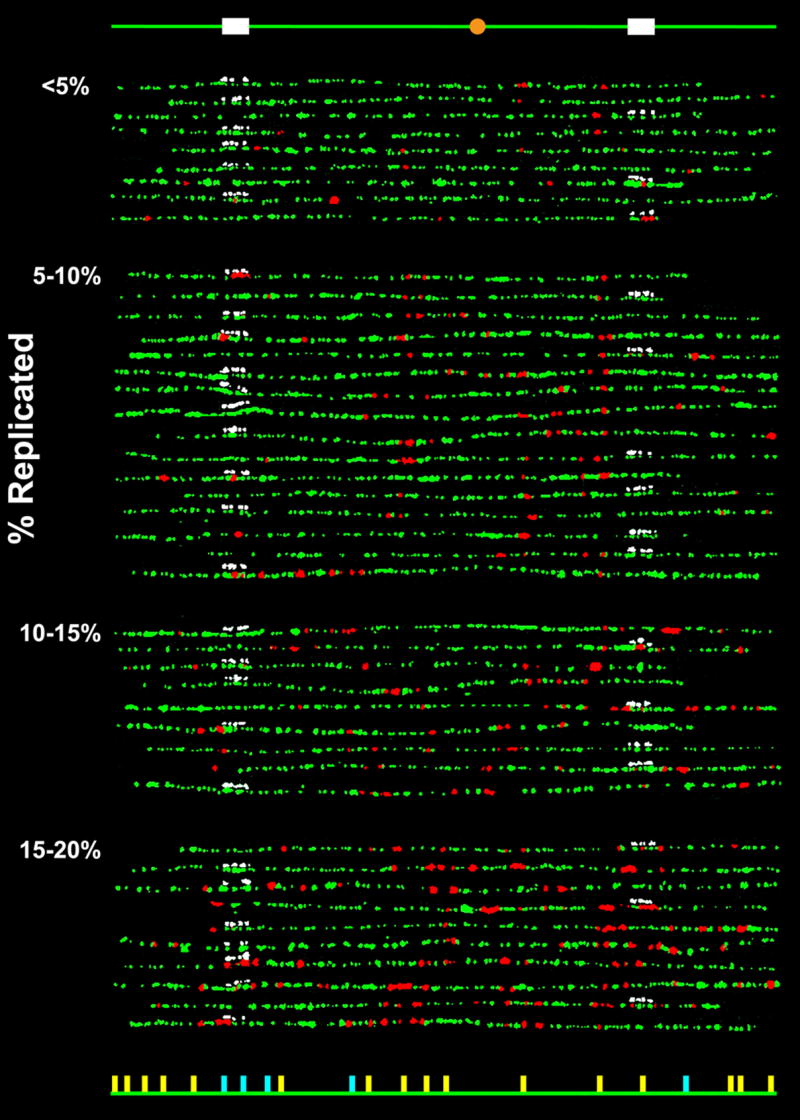
Replication patterns of individual chromosome VI molecules from α-factor synchronized budding yeast cells pulse-labeled during early S-phase with BrdU. Combed molecules were labeled by fluorophore-conjugated antibodies for BrdU (red) and DNA (green) as well as fluorophore-conjugated avidin (white) for the biotinylated hybridization probes to one of the two 10kb regions. The molecules were grouped according to their replication extent (indicated on the left). The schematic diagram at the top indicates the locations along the chromosome of the probes (white) and the centromere (orange), and the diagram at the bottom indicates the locations of the “confirmed” (yellow) and “potential” (light blue) origins. An origin is designated as “confirmed” if it has been identified in more than one of the previous mapping studies10; 11–13; 34–37 and “potential” if it has been identified only in a single study. It should be noted that there appears to be evidence in these patterns for previously unidentified origins, but most of these correlate with the locations of motifs found to be associated with origins36,37. A detailed analysis of this aspect of the data will be provided in a subsequent publication. The complete dataset is shown in Supplementary Fig. S2.
The strain used here (provided by K. Nasmyth) is K5409 (MATa ho ade2-1 trp1-1 can1-100 leu2-3, 112 his 3-11,15 ssd1 clb1-4ts ura3::URA3/GPD-TK).22 Cells were grown at 25°C in YPD to an OD600 of 0.5 and then incubated with α-factor (8μg/ml, GenScript) for 2.5hr. BrdU (Sigma) was added to a final concentration of 0.4mg/ml and 10 min later, the cells were released from the α-factor block with the addition of pronase (50μg/ml, Sigma). The cells were then washed with YPD 5 min after α-factor release to remove BrdU and then 30 min later, nocodazole (10μg/ml, Sigma) was added to arrest the cells, with fully replicated chromosomes, at the G2/M phase 2.25hr later. Nocodazole-arrested cells were recovered by centrifugation, resuspended in 10mM EDTA (pH 8.0), pelleted, and then resuspended in sorbitol buffer (1.0M sorbitol, 0.1M EDTA, 50mM DTT, pH 7.5) containing 0.7mg/ml lyticase at 37°C for 1hr. The cells were then pelleted, resuspended in sorbitol buffer, and mixed with an equal volume of 1.5% low-melting agarose (Bio-Rad) and cooled until the agarose had solidified, after which the plugs were incubated overnight at 37°C in 1mg/ml proteinase K (Bioline), 1% N-lauroylsarcosine, 0.5M EDTA, pH 9. Each plug contained roughly 108 cells. Pulse-field gel electrophoresis was performed in a 1% low-melting agarose gel. The band containing chromosome VI was excised, digested with β-agarase (New England Biolabs), and the DNA solution was then dialyzed overnight at 4°C in 5mM Tris, 1mM EDTA, pH8.0, using 100,000-molecular-weight-cutoff dialysis membrane (Spectrum). The solution was then adjusted to 0.2M MES, 5mM EDTA, pH 5.5 for DNA combing. To avoid DNA breakage, the purified DNA solution was handled with care to minimize shear. The glass slides onto which DNA molecules were combed were coated with octadecyltrichlorosilane (Sigma).38 The combed DNA molecules were denatured in 1M NaOH for 30min at room temperature, and hybridization was performed in 50% formamide, 1xSSC, 0.005% Tween 20, for 2hrs in the presence of 1μg/ml sheared salmonsperm DNA, 4μg/ml poly-uridine, 3μg/ml biotinylated probes at 37°C. The probe templates, prepared with PCR, covered one of two 10kb regions (only one of the regions, but not both, is used on each slide, except for distance calibration), in three blocks of ~2kb, 3kb, and 5kb: 45013–47023, 47051–52012, 52016–55231 and 210748–212827, 212915–217877, 217897–220277. The biotinylated probes were generated by random priming using BioPrime (Invitrogen). After incubation with Roche Blocking solution (Roche Biochemicals) for 30min, an alternating series of 45min incubations with (1/50 dilution) Alexa Fluor 350-neutrAvidin (Invitrogen) and (1/100 dilution) biotinylated anti-avidin antibodies (Vector Labs) labeled the probes. In total, there were 4 incubations with neutrAvidin and 3 incubations with biotinylated anti-avidin antibodies. The final set of incubations was: (1/50 dilution) anti-BrdU IgG (BU1/75 rat monoclonal, Abcam) for 1.5hrs, (1/70 dilution) Alexa Fluor 594-anti-rat IgG (rabbit, Invitrogen) for 1hr, (1/12 dilution) anti-ssDNA IgG (mouse, Argene) for 3hrs and then (1/30 dilution) Alexa Fluor 488-anti-mouse IgG (chicken, Invitrogen) for 1hr. All were carried out at room temperature. After all incubations, the sample was thoroughly washed with PBS. The sample was then fixed with 2% glutaraldehyde for 10min, and imaged in an anti fade solution (5.5% DABCO/45% glycerol (PBS)). Images were acquired using a Zeiss Axiovert S100 microscope equipped with a QImaging Retiga 1350EX CCD camera (25°C below ambient) and a 100× (NA=1.3) objective. The images were processed with Photoshop (Adobe), applying the same contrast/brightness adjustments (to the whole image) for all same-colored images. DNA lengths were calibrated using lambda phage DNA and chromosome VI molecules in which probes to both 10kb regions were used (data not shown).
