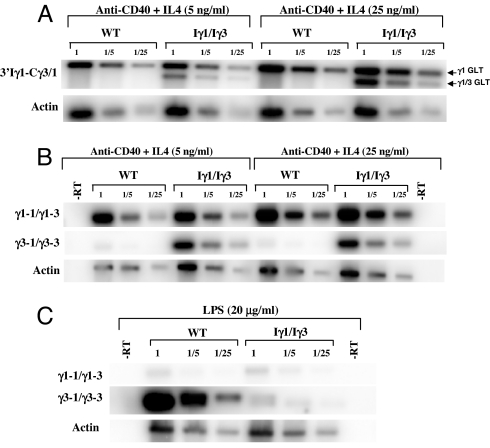
Fig. 4.
Quantification of GL transcripts. (A) Total RNA (day 3) from anti-CD40+IL4-activated WT or Iγ1/Iγ3 splenocytes was reverse-transcribed and the corresponding single-stranded cDNAs or dilutions thereof (1/5 and 1/25) were subjected to PCR, using 3′Iγ1-Cγ3/1 or actin primers. For quantification of the signals, a Southern blot analysis was performed. The nylon membranes were hybridized to 5′ end-radiolabeled Cγ3/1 or actin probes. Hybridization signals were quantified by a PhosphorImager. The ratios of 3′Iγ1-Cγ3/1 signals were corrected to the corresponding actin ratios. Arrows indicate transcripts initiating from the endogenous Iγ1 promoter (upper band, γ1 GLT) or from the replacement promoter (lower band, γ1/γ3 GLT). (B) For quantification of unspliced transcripts, nuclear RNA from of anti-CD40+IL4-activated WT or Iγ1/Iγ3 splenocytes was reverse-transcribed, and the corresponding single-stranded cDNAs or dilutions thereof (1/5 and 1/25) were subjected to PCR, using intronic primer pairs. For the signals' quantification, a Southern blot analysis was performed. The nylon membranes were hybridized to randomly primed intronic probes. Hybridization signals were quantified and corrected as in A. (C) Unspliced transcripts from LPS-activated WT or Iγ1/Iγ3 splenocytes were reverse-transcribed, and the corresponding single-stranded cDNAs or dilutions thereof (1/5 and 1/25) were subjected to PCR, using intronic primer pairs. The signals' quantification was performed as in A.