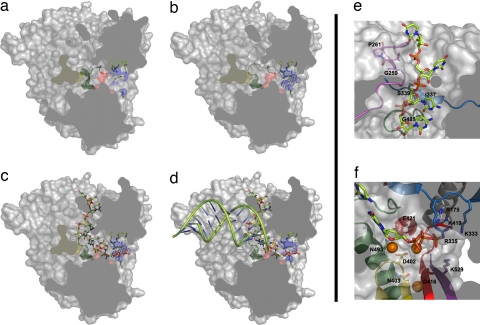
Fig. 4.
A model for activation of the VP1 polymerase and RNA synthesis. A space-filling representation of the enzyme (gray) has been cut to expose the three channels by which the different substrates accede to the active site (red surface). (a) In the absence of VP3, the B loop (green ribbon) partially occludes the binding site of the template base. (b) VP3 accedes to the VP1 active site, through the nucleotide entry tunnel, promoting the conformational change of the B loop that opens the catalytic cavity for template entry. Side chains of basic residues forming the tunnel are shown as sticks and blue surfaces. The electron density corresponding to the visible part of the VP3 peptide is shown as a blue chicken wire. (c) Once the template acceptor base reaches the active site, a second rearrangement should occur, facilitating the exit of VP3 and the entry of rNTP substrates to the active site. (d) After the catalysis of the first phosphodiester bond and pyrophosphate release, the newly synthesized RNA can ratchet down, displacing the C-terminal region (yellow surface). This movement would allow the exit of the nascent duplex out of the polymerase active site. (e) Model of VP1–RNA interactions. The motif G (pink) forms the entry of the template channel. The position of the template acceptor base in the VP1 active site is stabilized by interactions mediated by residues Ile-337 and Ser-339 of motif F (blue) in fingers and Gly-485 of motif B (green) in the palm. (f) The position of the incoming nucleotide appears stabilized by residues Asp-402, Asn-403 (motif C; yellow), Arg-529 (motif D; purple), Lys-333, and Arg-335 (motif F; blue), and residue Arg-175 (helix α5; gray). The docking model also shows how the two Mg2+ ions (orange balls) that coordinate the acidic residues Asp-402 (yellow) and Asp-416 and Asp-421 (red) of the active site are in good position to bind the incoming rNTP.