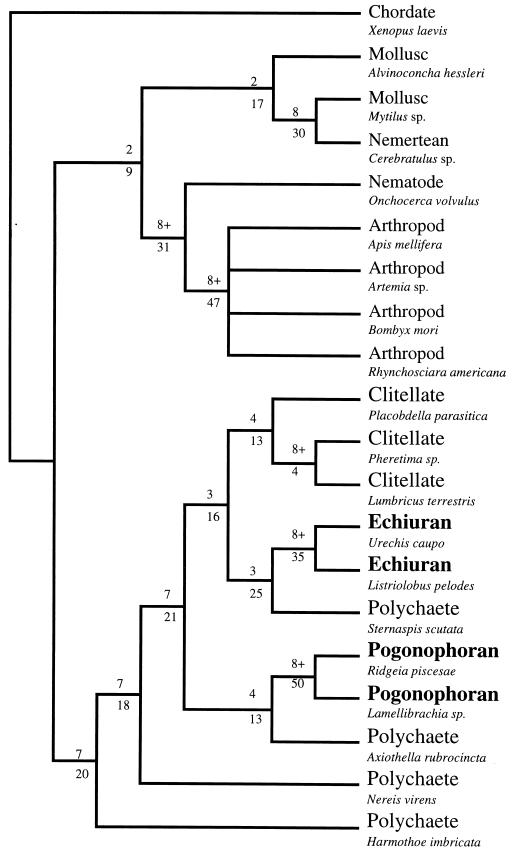
Figure 1.
Results of the phylogenetic analysis showing that echiurans and pogonophorans are derived groups within the annelid clade. The tree shown is the strict consensus of two equally parsimonious trees obtained with a paup (26) heuristic search (100 replicates, random addition search option) on the EF-1α nucleotide sequence data for 20 taxa (first and second codon positions only; 86 parsimony informative sites; length = 993; consistency index = 0.525; retention index = 0.522). The skewness test statistic for the distribution of tree lengths (27), g1 = −0.53, is significant (P < 0.01). Decay indices and branch lengths are shown above and below each node, respectively. The analysis shown incorporated a 4:1 transition/transversion ratio; ratios ranging from 2:1 to 10:1 yielded the same results. The deuterostome, Xenopus laevis, was designated as the outgroup for this analysis. If molluscs and arthropods are used as outgroups, the same worm clade topology results from maximum parsimony analysis, and neighbor joining analysis using the Kimura two-parameter distance model (28) with a γ correction for among-site rate variation also places echiurans and pogonophorans within a monophyletic worm clade.