Figure 10.
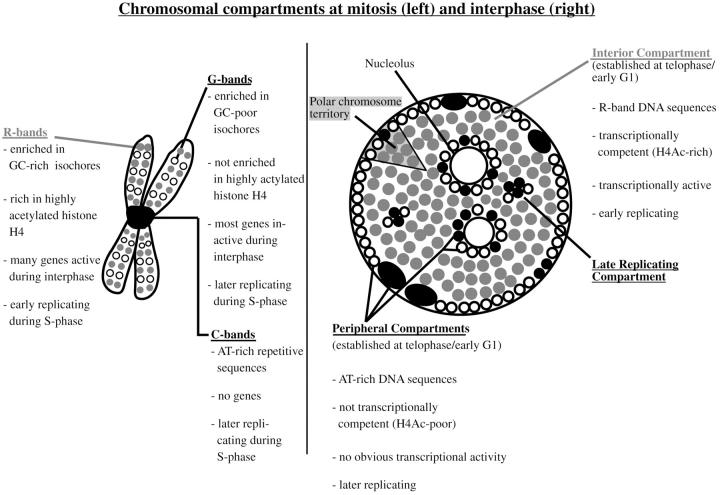
The scheme summarizes previous results and the data of the present paper regarding compartmentalization of mammalian genomes during mitosis and interphase. The well characterized bands of mitotic chromosomes give rise to distinct higher order functional compartments within the cell nucleus. Distinct bands of mitotic chromosomes differ in a variety of features as isochore composition and corresponding DNA sequence composition (Bickmore and Sumner 1989; Craig and Bickmore 1993; Bernardi 1995), gene content (Bickmore and Sumner 1989; Craig and Bickmore 1993; Bernardi 1995; Cross et al. 1997), acetylation levels of histone H4 (Jeppesen and Turner 1993), transcriptional activity of genes (Craig and Bickmore 1993, Craig and Bickmore 1994), and replication timing during interphase (Dutrillaux et al. 1976; Camargo and Cervenka 1982). Differences in DNA sequence composition (Rae and Franke 1972; Manuelidis and Borden 1988; O'Keefe et al. 1992), acetylation levels of histone H4 (acetylated histone H4 designated as H4Ac), transcriptional activity, and replication timing of chromatin targeted to distinct nuclear compartments (Ferreira et al. 1997) demonstrate the functional features of these compartments and their relation to genome organization revealed by banding patterns of mitotic chromosomes. R-band sequences (symbolized by gray dots) localize to the interior compartment, whereas G- and C-band sequences localize to the peripheral and late replicating compartments (symbolized by black and open dots). In the present study, the peripheral and late replicating compartments revealed comparable properties. Higher order nuclear compartments are built up by chromosome territories displaying a distinct polarized distribution of R-band DNA and G/C-band DNA organized into corresponding subchromosomal foci (Zink et al. 1998, Zink et al. 1999).