Figure 8.
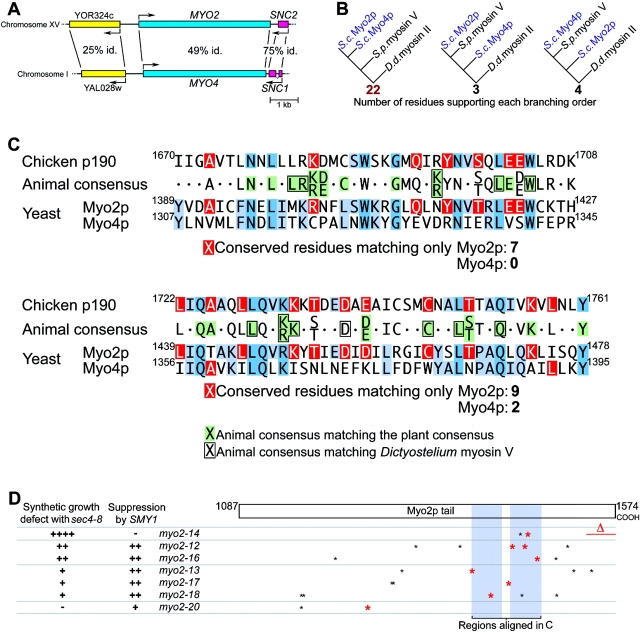
Myo2p has features conserved with animal myosin Vs but lost from Myo4p. (A) Map of genomic regions near MYO2 and MYO4. Percentage of amino acid identity is shown (P ≤ 10−21 for all three BLASTP 2.0 scores). (B) Evolutionary relationship among the motor domains of Myo2p, Myo4p, and their closest relative, Schizosaccharomyces myosin V (CAA22641), using Dictyostelium myosin II as an outgroup. 22/29 corresponds to P(χ2) ≤ 10−6 and a bootstrap value of 99.7%. (C) Comparison of unambiguously alignable regions of p190, Myo2p, and Myo4p, showing consensus sequence for animal myosinVs, and matches of the animal consensus with the plant consensus (green) and Dictyostelium myosin V (boxed) sequences. Residues in the consensus sequence matching only Myo2p or only Myo4p are shown in red and other matches among the sequences are shown in blue. The animal consensus represents residues that match in at least six of the seven animal myosin Vs. (D) Summary of genetic interactions and positions of myo2ts mutations relative to the regions aligned in C. Nonconservative mutations in residues that are conserved with animals are shown in red.