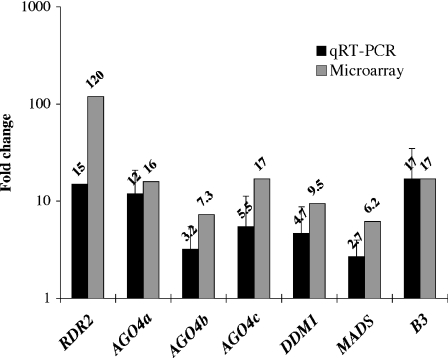
Figure 3.
Quantitative RT-PCR (qRT-PCR) analyses on seven significantly up-regulated expressed sequence tags (ESTs) derived from the microarray analysis. Fold changes [shoot apical meristem (SAM)/seedling] from qRT-PCR analyses (black bars) and the microarray analysis (gray bars) are indicated on a logarithmic scale with seven significantly up-regulated ESTs derived from the microarray analysis: RDR2, AGO4a (accession no. DV493575), AGO4b (accession no. DV943274), AGO4c (accession no. DV492375), DDM1 (Table 3), MADS, and B3. AGO4a, AGO4b, and AGO4c have similar but distinct sequences (data not shown), indicating that these ESTs were derived from paralogous loci (Table 3). Means of the two biological replications (RDR2) and means + SD of the three biological replications (AGO4a, AGO4b, AGO4c, DDM, MADS, and B3) are shown. For Cinful and Tekay, one of the seedling samples (replication 4, Appendix S1) did not yield fluorescence above the threshold level whereas the corresponding SAM sample in the same replication did yield fluorescence above threshold level. Fold changes of the single replications for Cinful and Tekay were 7904 and 446, respectively.