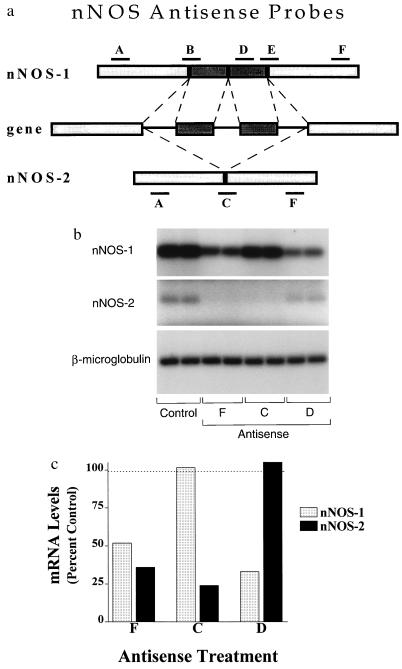
Figure 1.
Effect of antisense probes against nNOS mRNA. (a) A schematic of a portion of the nNOS gene is presented which illustrates both nNOS-1 and nNOS-2 as splice variants. Antisense A and F are present on both isoforms. Antisense B and E span the splice regions between the conserved and the additional exons in nNOS-1. Antisense D targets one of the additional exons in nNOS-1. Antisense C spans the splice site where the two exons in nNOS-1 are deleted and is selective for nNOS-2. The sequences of the oligodeoxynucleotides are as follows: antisense A, GGG CTG GGT CAC ACG GAT GG(nt 454–474); antisense B, GCT GTA TAC AGA TCT CTG TGA ACT CC (nt 1595–1621); antisense C, TCT TGG CTA CTT CCT GTG TGA ACT CC (nt 1595–1608, 1922–1935); antisense D, CGA ACG CCA ATC TCC GTG CC (nt 1866–1886); antisense E, TCT TGG CTA CTT CCT CCA GGA TGT TG (nt 1910–1936); antisense F, GAA TCC TCT CCC CGC CCA (nt 2815–2833); mismatch C, TCT GTG CTA CTC TCT CTG TGA ACT CC; mismatch D, CAG ACG CAC ATC CTC GTG CC; mismatch F, GAA TCT CCT CCC GCC CCA (from GenBank accession no. D14552). These correspond to the following exons based upon the human gene: A, exon 2; B, exon 8/9; C, exon 8/11; D, exon 10; E, exon 10/11; F, exon 18. (b) The levels of nNOS-1, nNOS-2, and β2-microglobulin were determined in duplicate control and antisense F treated samples using RT-PCR as described. (c) The corresponding bands observed on the film were cut out and counted. The two samples were averaged and plotted as percent of control.